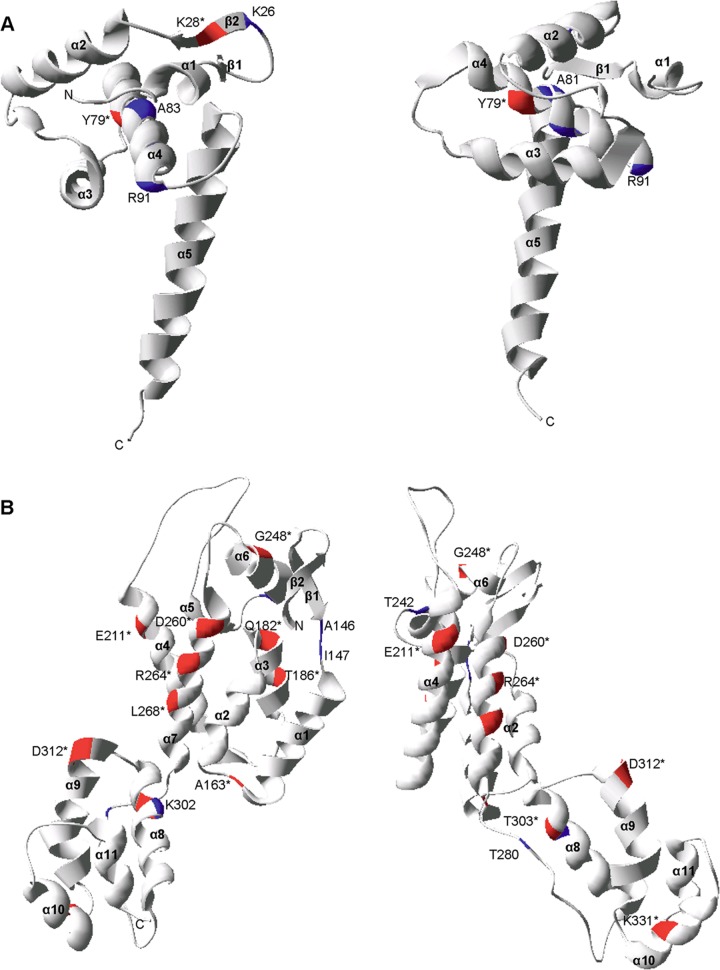
Fig 4.
Structural locations of the CTL escape mutations in HIV-1 Gag p17 and p24. The CTL escape mutations mapped onto two rotations of the ribbon representation of the three-dimensional monomer structures of p17 (PDB: 1HIW) (A) and p24 (PDB: 3GV2) (B). The locations of CTL escape mutations are denoted by the consensus amino acid and the Gag amino acid position. Mutations that caused a statistically significant impact on virus replication are denoted by an asterisk and by red highlighting of the ribbon. All other mutations are denoted by blue highlighting.