Abstract
Toscana virus (TOSV), which is transmitted by Phlebotomus spp. sandflies, is a major etiologic agent of aseptic meningitis and encephalitis in the Mediterranean. Like other members of the genus Phlebovirus of the family Bunyaviridae, TOSV encodes a nonstructural protein (NSs) in its small RNA segment. Although the NSs of Rift Valley fever virus (RVFV) has been identified as an important virulence factor, which suppresses host general transcription, inhibits transcription from the beta interferon promoter, and promotes the proteasomal degradation of double-stranded RNA-dependent protein kinase (PKR), little is known about the functions of NSs proteins encoded by less-pathogenic members of this genus. In this study we report that TOSV is able to downregulate PKR with similar efficiency as RVFV, while infection with the other phleboviruses—i.e., Punta Toro virus, sandfly fever Sicilian virus, or Frijoles virus—has no effect on cellular PKR levels. In contrast to RVFV, however, cellular transcription remains unaffected during TOSV infection. TOSV NSs protein promotes the proteasome-dependent downregulation of PKR and is able to interact with kinase-inactive PKR in infected cells.
INTRODUCTION
The family Bunyaviridae includes the animal-infecting genera Orthobunyavirus, Phlebovirus, Hantavirus, and Nairovirus and the plant-infecting genus Tospovirus. Phleboviruses are comprised of the sandfly fever group (which is vectored by mosquitoes or phlebotomine sandfiles) and the Uukuniemi group (which is vectored by ticks). The sandfly group consists of the Punta Toro serocomplex (e.g., Punta Toro virus [PTV]), the Naples serocomplex (e.g., Toscana virus [TOSV]), the Icoaraci serocomplex, the Frijoles serocomplex (e.g., Frijoles virus [FRIV]), the Sicilian serocomplex (e.g., sandfly fever Sicilian virus [SFSV]) as well as the serologically distinct Rift Valley fever virus (RVFV) (1). Several members of this group—including RVFV, TOSV, SFSV, and PTV—are pathogenic to humans and are of important public health concern. TOSV is an enveloped virus carrying a negative-sense, single-stranded RNA genome comprising three segments, which are designated L (large), M (medium), and S (small). The L-segment encodes the viral polymerase (L), the M-segment encodes the envelope glycoproteins Gn and Gc, and the S-segment encodes the nucleoprotein (N) (2). In addition, the S-segment also encodes the nonstructural protein NSs in an ambisense manner (3).
TOSV is an emerging pathogen and the major etiologic agent of aseptic meningitis and encephalitis in the Mediterranean (4–7). TOSV is transmitted by sandflies (Phlebotomus perniciosus and Phlebotomus perfiliewi), and the incidence of TOSV infection peaks during the summer months when its insect vectors are most active (8). No animal reservoir of TOSV has been identified to this date, although its insect vector may also act as a reservoir, since both transovarial and venereal transmission of TOSV between Phlebotomus spp. sandflies has been observed (4). It is not known whether humans have the potential to act as amplifying host during an outbreak.
Phlebotomous fever caused by sandfly-borne phleboviruses is characterized by self-limiting febrile illness in humans (8). On the other hand, the mosquito-borne RVFV is a zoonotic virus that causes hemorrhagic fever, neurological disorders, or blindness in humans and leads to high rates of abortion or fetal malformation in pregnant ruminants, as well as high mortality in newborn lambs (9). Among the sandfly-borne phleboviruses, TOSV has unique pathological manifestations and often causes disease with central nervous system (CNS) involvement. The most frequently observed symptoms of TOSV infection in humans are aseptic meningitis, meningoencephalitis, and encephalitis, as well as less severe manifestations without CNS involvement, such as febrile erythema and generalized influenza-like illness. In addition, an increasing number of unusual clinical manifestations and severe sequelae, such as hydrocephalus and ischemic complications, have been reported during recent years. Seroprevalence studies in the regions where TOSV is endemic suggest a large number of asymptomatic infections (10). There are no vaccines or effective treatment measures available for TOSV infection, and the pathological mechanism remains unclear.
Phlebovirus NSs proteins play an important role in viral evasion from host innate immune responses. The NSs protein of RVFV has been the most thoroughly characterized to date and employs at least three independent mechanisms to subvert host cells defenses: it inhibits the activation of the beta interferon (IFN-β) promoter through interaction with the repressor protein SAP30 (11), it effects a generalized suppression of host cell transcription by sequestering the TFIIH subunit p44 (12) and promoting the proteasomal degradation of the TFIIH subunit p62 (13), and, in addition, it is able to target a second protein, double-stranded RNA-dependent protein kinase (PKR), for proteasomal degradation (14, 15). Furthermore, RVFV NSs interacts with pericentromeric DNA sequences through its SAP30-binding domain (16) and induces a DNA damage signaling response (17). Considerably less effort has thus far been expended to study the NSs proteins of other phleboviruses. However, it is known that SFSV (14) and PTV (18) NSs inhibit the upregulation of IFN-β and that SFSV NSs does not promote the degradation of PKR (14). Furthermore, TOSV NSs has recently been shown to suppress IFN-β induction by inhibiting the dimerization of IRF-3 (19).
PKR mediates a critical role in the cellular host defense by acting as a sensor of viral infection. PKR binds to double-stranded RNA or 5′-triphosphated single-stranded RNA at the N-terminal RNA-binding domain, which causes homodimerization, and exposes the C-terminal serine/threonine kinase domain leading to autophosphorylation of the kinase domain. Activated PKR then phosphorylates eukaryotic initiation factor 2α (eIF2α). eIF2 with the phosphorylated α subunit binds to eIF2B with high affinity and prevents the eIF2B-mediated exchange of eIF2-GDP into eIF2-GTP, which leads to the inhibition of translation initiation (20). RVFV NSs promotes degradation of PKR and prevents the shutoff of viral translation (14, 15).
Since TOSV causes a unique pathology with CNS involvement among the phlebotomus fevers, we hypothesized that TOSV NSs encodes another virulence function in addition to IFN-β suppression. In the present study, we therefore aimed to identify novel functions of TOSV NSs. As a result, we found that TOSV is able to promote the degradation of PKR but is unable to suppress host general transcription. TOSV NSs downregulates PKR with similar kinetics and efficiency as RVFV NSs. TOSV NSs protein is able to bind to kinase-inactive PKR in infected cells and promotes the proteasomal degradation of PKR. The characterization of this novel TOSV NSs function will be important for understanding the pathology of TOSV infection in humans.
MATERIALS AND METHODS
Cells and viruses.
293 and VeroE6 cells were maintained in Dulbecco modified minimum essential medium supplemented with 10% fetal bovine serum (FBS) and 100 μg of penicillin-streptomycin/ml (all from Invitrogen). BHK/T7-9 cells (21), which stably express T7 RNA polymerase, were grown in MEM-α supplemented with 10% FBS and 100 μg of penicillin-streptomycin/ml (all from Invitrogen) and 600 μg of hygromycin (Cellgro)/ml. The RVFV vaccine candidate MP-12 (22), as well as all recombinant MP-12 mutants, were amplified in VeroE6 cells, and the infectivities were determined by plaque assay in the same cells. TOSV (ISS.Phl.3), PTV (D-4021A), FRIV (VP-161A), and SFSV (Sabin) were obtained from R. B. Tesh at the World Reference Center for Emerging Viruses and Arboviruses at the University of Texas Medical Branch and passaged in VeroE6 cells up to two times.
Analysis of virus replication.
VeroE6 cells were infected with the indicated viruses at a multiplicity of infection (MOI) of 1, and cell culture supernatants were harvested at 0, 12, 24, 48, 72, and 96 h postinfection (hpi). Plaque assays were performed as previously described (23, 24). Plaques of MP-12 were imaged at 4 days postinfection (dpi), those of TOSV and FRIV were imaged at 7 dpi, and those of PTV and SFSV were imaged at 8 dpi, respectively.
Antibodies and reagents.
Anti-myc (9E10), anti-GAPDH (V-18), anti-β-actin (I-19), and the horseradish peroxidase (HRP)-labeled anti-mouse, anti-goat and anti-rabbit secondary antibodies were purchased from Santa Cruz. The conformation-specific anti-rabbit-HRP antibody (L27A9) used to detect TOSV NSs after immunoprecipitation was purchased from Cell Signaling. Anti-FLAG M2 (F3165) and anti-GFP (G1544) antibodies were purchased from Sigma. The anti-human PKR (610764) monoclonal antibody was purchased from BD Biosciences. The antisera against RVFV, TOSV, PTV, FRIV, and SFSV were a gift from R. B. Tesh at the University of Texas Medical Branch. Goat anti-mouse IgG (H+L, Alexa Fluor 488 coupled), goat anti-rabbit (H+L, Alexa Fluor 488 coupled), and donkey anti-goat (H+L, Alexa Fluor 594 coupled) antibodies were purchased from Invitrogen. The anti-TOSV NSs rabbit polyclonal antibody (TOSNS302) was generated by immunizing rabbits with the C terminus (residues 302 to 316, VTKWLPSPPHYPPL) of the TOSV NSs protein (EZBiolab). Leptomycin B and chloroquine were purchased from Santa Cruz, and lactacystin was purchased from Enzo Life Sciences. Actinomycin D (ActD) and MG132 were purchased from Sigma. Cycloheximide (CHX) was purchased from Acros Organics.
Virus rescue.
The recombinant MP-12 mutants, rMP12-TOSNSs and rMP12-TOSNSs-Flag were recovered as previously described (23). Briefly, subconfluent monolayers of BHK/T7-9 cells were cotransfected with a mixture of the respective pProT7-S(+) plasmid (see below), pPro-T7-M(+), pPro-T7-L(+), pT7-IRES-vN, pCAGGS-vG, and pT7-IRES-vL using TransIT-LT1 (Mirus Bio Corp.) according to the manufacturer's instructions. The culture medium was replaced with fresh medium 24 h later. At 5 days posttransfection, the culture supernatants were collected, clarified, and then inoculated into fresh VeroE6 cells. The culture supernatant from VeroE6 cells was collected at 3 dpi, and the virus titer was determined by plaque assay. The RVFV NSs truncation mutant rMP12-C13type has been described previously (23).
Plasmids.
pProT7-S(+)-TOSNSs and pProT7-S(+)-TOSNSs-Flag were generated by PCR amplification of the TOSV NSs ORF using the following primer pairs (HpaTOSF [AGT TGT TAA CAT GCA ATC CAG AGC TGT CAT CTT G] and SpeTOSR [CGC TAC TAG TTC ATA AGG GTG GGT AGT GGG GGG GAC] or HpaTOSF and SpeFlagTOSR [CGC TAC TAG TCT ACT TAT CGT CGT CAT CCT TGT AAT CTA AGG GTG GGT AGT GG]), respectively. The resulting PCR fragment was then cloned between the HpaI and SpeI sites of pProT7-S(+). pcDNA3.1mycHisA-TOSNSs was generated as follows. PCR fragment (KpnTOSNSF [AGT TGG TAC CAT GCA ATC CAG AGC TGT CAT C] and XhoTOSNSR [AGT TCT CGA GTC ATA AGG GTG GGT AGT GG]) was digested with KpnI and XhoI and cloned into pcDNA3.1-mycHisA (Invitrogen). pCAGGS-GFP-PKRK296R was generated as follows. PCR fragment encoding the green fluorescent protein (GFP) ORF (ClaGFPF [ATG GAT CGA TAT GGT GAG CAA GGG CGA GGA GCT GTT CAC C] and NotAatGFPR [TCC TCC TCC CGC GGC CGC GAC GTC CTT GTA CAG CTC GTC CAT GCC GAG AGT G]) was inserted into the pCAGGS plasmid between the ClaI and NotI sites and designated as the pCAGGS-GFP cassette. The PCR fragment encoding PKRK296R (PKR1F [GTC GCG GCC GCG GGA GGA GGA ATG GCT GGT GAT CTT TCA GCA GGT TTC TTC] and PKR5R [ATG GCT CGA GCT AAC ATG TGT GTC GTT CAT T]) was then inserted between the NotI and XhoI sites of pCAGGS-GFP cassette. pT7-IRES-GFP has been described previously (25). pcDNA3.1myc-PKRK296R and pcDNA3.1myc-CAT were described previously (15).
RNA synthesis and transfection.
Capped and polyadenylated RNA transcripts were synthesized in vitro from pcDNA3.1mycHisA-TOSNSs and pT7-IRES-GFP by using the mMESSAGE mMACHINE T7 Ultra kit (Ambion) according to the manufacturer's instructions. A 1-μg portion of in vitro-synthesized RNA was transfected into 293 cells in a 12-well plate using a TransIT-mRNA transfection kit (Mirus Bio Corp.) according to the manufacturer's instructions.
Quantitative real-time PCR.
293 cells were mock infected or infected with MP-12 at an MOI of 3. At 8 h posttransfection, the cells were harvested with a rubber policeman and washed once in phosphate-buffered saline (PBS), and the total RNA was purified from 6 × 105 cells using an RNeasy minikit (Qiagen) according to the manufacturer's instructions. First-strand cDNA was synthesized from 1 μg of RNA using a high-capacity cDNA reverse transcription kit (Applied Biosciences). Real-time PCR was performed on a Mastercycler ep realplex2 (Eppendorf) using a QuantiFast SYBR green PCR kit and QuantiTect primer assays QT01883280 (Hs_EIF2AK2_2_SG [PKR]) and QT01192646 (glyceraldehyde 3-phosphate dehydrogenase [GAPDH]) (all from Qiagen) according to the manufacturer's instructions.
Western blot analysis.
Western blot analysis was performed as previously described (15). Briefly, samples were normalized by cell number and disrupted by boiling in sample buffer for 10 min, followed by SDS-PAGE. The proteins were then transferred onto Immobilon P polyvinylidene fluoride membranes (Millipore) and probed with the indicated antibodies.
Click chemistry and immunofluorescence.
Labeling of nascent RNA was performed as previously described (13). Briefly, cells grown on 12-mm coverslips were incubated in medium containing 1 mM 5-ethynyl uridine (5-EU; Berry & Associates) for 1 h prior to fixation in 4% paraformaldehyde. The cells were then permeabilized in 0.2% Triton X-100, and nascent RNA with incorporated 5-EU was detected by click chemistry using 20 μM Alexa Fluor 594-azide (Invitrogen). The cells were mounted on coverslips using Fluoromount-G (Southern Biotech), and images were acquired on an Olympus IX71 fluorescence microscope using a ×20 LCPlanFl (NA 0.4) objective lens.
FACS analysis.
Fluorescence-activated cell sorting (FACS) analysis was carried out as previously described (13). Briefly, 293 cells were mock infected or infected with TOSV at an MOI of 3 and treated with 0.5 mM 5-EU for 1 h prior to harvesting. Some samples were treated with 5 μg of ActD/ml concurrently with the 5-EU treatment to suppress cellular RNA synthesis. After harvesting, the cells were fixed in 4% paraformaldehyde for 30 min and permeabilized in 0.2% Triton X-100 in PBS for 10 min, and nascent RNA with incorporated 5-EU was detected by click chemistry using 200 nM Alexa Fluor 647-azide (Invitrogen). The cells were then stained with a mouse polyclonal antibody against TOSV, followed by an Alexa Fluor 488-labeled secondary antibody, to detect the expression of viral proteins. The cells were analyzed by flow cytometry on an LSRII Fortessa instrument (BD Biosciences).
Coimmunoprecipitation.
To precipitate Flag-tagged TOSV NSs with PKR-GFP, cells were transfected with pCAGGS-GFP-PKRK296R and subsequently infected with rMP12-TOSNSs-Flag at an MOI of 3 at 12 h posttransfection. The cells were harvested at 12 hpi by scraping them with a rubber policeman and lysis, and immunoprecipitation with magnetic beads conjugated with anti-GFP antibody was performed using the μMACS GFP tag protein isolation kit (catalog no. 130-091-125; Miltenyi Biotech) according to the manufacturer's instructions. To precipitate untagged TOSV NSs with PKR-GFP, 293 cells were transfected with pCAGGS-GFP-PKRK296R and subsequently infected with TOSV at an MOI of 3 at 16 h posttransfection. The cells were harvested at 8 hpi, and lysis and immunoprecipitation were performed as described above. To precipitate myc-PKR with untagged TOSV NSs, the cells were transfected with pcDNA3.1myc-PKRK296R and subsequently infected with TOSV at an MOI of 3 at 16 h posttransfection. The cells were harvested at 8 hpi, and lysis and immunoprecipitation with anti-TOSV NSs antibody (TOSNS302) and magnetic beads conjugated to protein G (Invitrogen) were performed as previously described (13). Cleared lysates and precipitates were then analyzed by Western blotting. The band intensities were quantified using QuantityOne software from Bio-Rad.
RESULTS
TOSV promotes the downregulation of PKR.
RVFV NSs promotes the degradation of PKR (14, 15), and PKR plays an important role in host defense against RVFV infection (14). Since SFSV NSs does not promote the degradation of PKR (14), we sought to determine whether other representative phleboviruses in the sandfly fever group (i.e., TOSV, PTV, and FRIV) (1) have the ability to downregulate PKR. SFSV was also included as a control that does not promote the degradation of PKR. We used the RVFV MP-12 vaccine strain (22), since its NSs protein is fully functional (13, 15, 23, 26) and differs from the virulent, parental ZH548 strain by only one, non-attenuating amino acid substitution (27). The rMP12-C13type is a recombinant virus encoding a 69% in-frame truncation of the NSs gene in the MP-12 backbone (23) and was used as a control RVFV lacking all NSs functions.
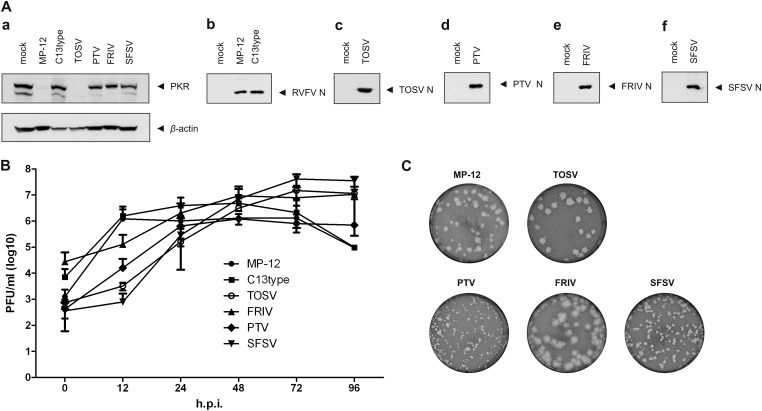
All viruses were amplified in VeroE6 cells and then used to infect VeroE6 cells at an MOI of 1. Since PKR is inducible by type I IFNs (28), VeroE6 cells, which lack IFN-α/β genes (29, 30), were chosen for this experiment to minimize possible effects of type I IFN-mediated PKR induction. Cells infected with MP-12, rMP12-C13type, TOSV, PTV, or FRIV were harvested at 24 hpi, while those infected with SFSV were collected at 48 hpi due to relatively slow replication kinetics in VeroE6 cells (Fig. 1B). Whole-cell lysates were then analyzed by Western blotting with anti-PKR antibodies to determine PKR expression levels (Fig. 1Aa). Whole-cell lysates were also probed with specific mouse polyclonal sera raised against each individual virus (Fig. 1Ab to f) to detect viral protein expression in the infected cells. As shown in Fig. 1A, both RVFV MP-12 and TOSV were able to downregulate PKR in infected cells, whereas PTV, SFSV, and FRIV NSs did not promote PKR downregulation in infected cells. All viruses replicated to titers between 1.62 × 105 (TOSV) and 7.01 × 106 (SFSV) at the times sampled in Fig. 1A, indicating that that the lack of PKR degradation by FRIV, PTV, and SFSV is not due to poor viral replication (Fig. 1B). TOSV, PTV, FRIV, and SFSV formed plaques distinct from those of MP-12 (Fig. 1C).
Fig 1.
RVFV and TOSV infection downregulate PKR expression. (A) VeroE6 cells were mock infected or infected with RVFV MP-12 strain (MP-12), rMP12-C13type (C13type), Toscana virus (TOSV), Punta Toro virus (PTV), Frijoles virus (FRIV), or sandfly fever Sicilian virus (SFSV) at an MOI of 1, and the cells were harvested at 48 hpi (SFSV) or 24 hpi (all others). Whole-cell lysates were analyzed by Western blotting with anti-PKR and anti-β-actin antibodies (a) or polyclonal antisera raised against RVFV (b), TOSV (c), PTV (d), FRIV (e), or SFSV (f), respectively. (B) VeroE6 cells were infected with MP-12 (●), C13type (■), TOSV (○), FRIV (▲), PTV (◆), or SFSV (▼) at an MOI of 1. Supernatants of infected cultures were harvested at the indicated time points postinfection, and virus titers were determined by plaque assay. Shown are the means and standard deviations (SD) of two independent experiments. (C) Plaque morphologies of MP-12, TOSV, PTV, FRIV, and SFSV in VeroE6 cells stained with neutral red.
TOSV does not suppress host cell transcription.
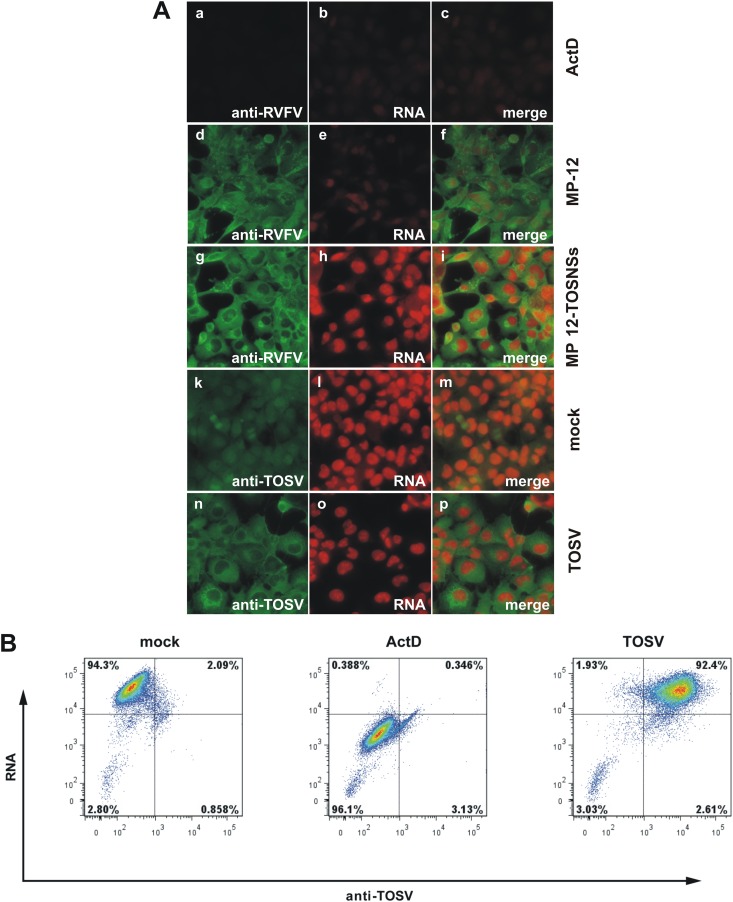
To further characterize the functional differences between TOSV and RVFV NSs, we next tested whether TOSV NSs could suppress host general transcription. To this aim, 293 cells were mock infected or infected with TOSV (MOI = 1), MP-12 (MOI = 3), or a recombinant MP-12 encoding TOSV NSs in place of MP-12 NSs (rMP12-TOSNSs, MOI = 3). At 23 hpi, the uridine analog 5-EU was added to the growth medium for 1 h, and the cells were fixed at 24 hpi. 5-EU is incorporated into nascent RNA instead of uridine and can be detected using “click” chemistry (31). We visualized newly synthesized RNA using an Alexa Fluor 594-coupled azide (Fig. 2A, shown in red) and expression of viral proteins by indirect immunofluorescent assay using anti-RVFV (Fig. 2Aa to i) or anti-TOSV (Fig. 2Ak to p) antisera, followed by a secondary Alexa Fluor 488-coupled antibody (shown in green). Mock-infected cells (Fig. 2Ak to m) served as the positive control for active RNA synthesis, whereas ActD-treated cells served as the control for transcriptional suppression (Fig. 2Aa to c). Consistent with our previous results (13), MP-12 was able to suppress cellular general transcription (Fig. 2Ad to f). Neither authentic TOSV (Fig. 2An to p) nor rMP12-TOSNSs (Fig. 2Ag to i) were able to suppress transcription in infected cells, even though all cells expressed abundant viral proteins, as shown by immunofluorescent staining with virus-specific antisera.
Fig 2.
TOSV NSs does not suppress cellular general transcription. (A) 293 cells were mock infected (a to c, k to m) or infected with TOSV at an MOI of 1 (n to p), RVFV MP-12 at an MOI of 3 (d to f), or rMP12-TOSNSs at an MOI of 3 (g to i). From 23 to 24 hpi, cells were treated with 1 mM 5-ethynyl uridine (5-EU) to label newly synthesized RNA. As a control for transcriptional suppression, cells were treated with 5 μg of ActD/ml concurrently with the 5-EU treatment (a to c). After fixing and permeabilization, labeled RNA was detected by click chemistry with an Alexa Fluor 594-coupled azide (red). To visualize expression of viral proteins, the cells were stained with either a polyclonal anti-RVFV (a to i) or anti-TOSV (k to p) antibody, followed by an Alexa Fluor 488-coupled secondary antibody (green). The cells were analyzed by fluorescence microscopy. (B) 293 cells were mock infected or infected with TOSV at an MOI of 1. From 14 to 16 hpi, the cells were treated with 0.5 mM 5-EU to label newly synthesized RNA. As a control for transcriptional suppression, cells were treated with 5 μg of ActD/ml concurrently with the 5-EU treatment. After fixing and permeabilization, labeled RNA was detected by click chemistry with an Alexa Fluor 647-coupled azide. To visualize expression of viral proteins, the cells were stained with a polyclonal anti-TOSV antibody, followed by an Alexa Fluor 488-coupled secondary antibody. The cells were analyzed by flow cytometry.
To obtain quantitative data on the effect of TOSV replication on host cell transcription, we performed flow cytometry (Fig. 2B). Again, 293 cells were infected with TOSV at an MOI of 1, and newly synthesized RNA was labeled using 5-EU from 14 to 16 hpi. As for the immunofluorescence described above, the cells were treated with ActD concurrently with the 5-EU labeling to serve as a control for transcriptional suppression. RNA was visualized using an Alexa Fluor 647-coupled azide, and TOSV protein expression was detected using anti-TOSV antibodies, followed by a secondary Alexa Fluor 488-coupled antibody. As shown in Fig. 2B, almost all cells were infected with TOSV, and the population of TOSV-infected cells had the same level of RNA fluorescence intensity as mock-infected cells (92.4 and 94.3%, respectively), while only 0.4% of the ActD-treated cells incorporated 5-EU. These results clearly demonstrate that TOSV, in contrast to RVFV, is not able to effect a host cell general transcriptional suppression.
TOSV NSs protein downregulates PKR early during infection.
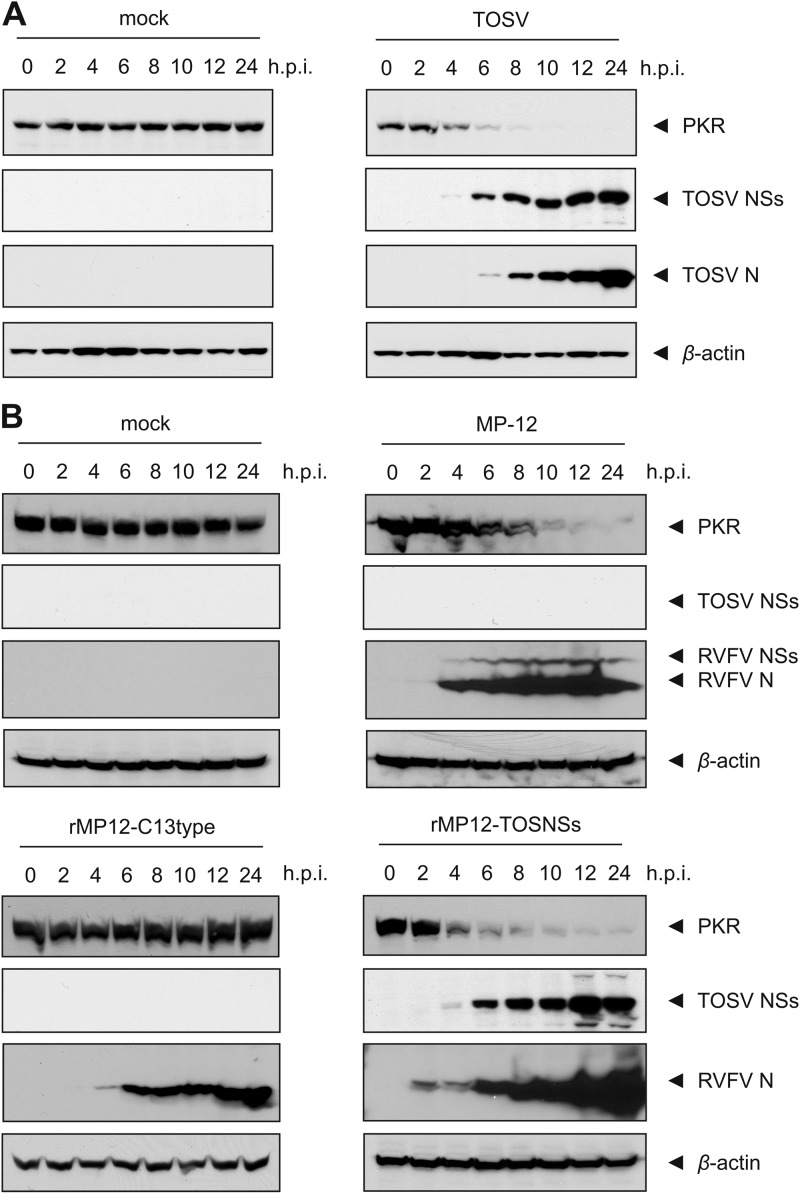
In order to elucidate the kinetics of TOSV-mediated PKR downregulation, as well as the expression of viral proteins, 293 cells were infected with TOSV at an MOI of 3 and harvested at 0, 2, 4, 6, 8, 10, 12, and 24 hpi. Whole-cell lysates of cells collected at the indicated time points were analyzed by Western blotting with anti-PKR, anti-TOSV NSs, and anti-TOSV antibodies (Fig. 3A). The beginning of PKR downregulation in TOSV-infected cells coincided with the expression of the NSs protein, which could be detected as early as 4 hpi and continued to increase in abundance until the last time point studied at 24 hpi. Expression of the N protein could also be detected as early as 4 hpi in longer exposures. The abundance of PKR was decreased to below detection levels between 8 and 10 hpi, whereas the expression of PKR in mock-infected cells remained unchanged over the 24-h period studied.
Fig 3.
Kinetics of PKR downregulation. (A) Time course of PKR downregulation during TOSV infection. 293 cells were infected with TOSV at an MOI of 3 (right panel) or mock infected (left panel). Cells were harvested at the indicated time points postinfection and whole-cell lysates were analyzed by Western blotting with anti-PKR, anti-TOSV NSs (TOSNS302), anti-TOSV, or anti-β-actin antibodies (from top to bottom), respectively. (B) Time course of PKR downregulation by TOSV and RVFV NSs. 293 cells were mock infected (top left panel) or infected with a recombinant RVFV that carries a 69% deletion of the NSs-ORF (rMP12-C13type, bottom left panel), RVFV MP-12 (top right panel), or a recombinant MP-12 in which the NSs-ORF has been replaced with that of TOSV NSs (rMP12-TOSNSs, bottom right panel) at an MOI of 3. The cells were harvested at the indicated time points postinfection, and whole-cell lysates were analyzed by Western blotting with anti-PKR, anti-TOSV NSs, anti-RVFV, or anti-β-actin antibodies (from top to bottom), respectively.
To compare the ability of TOSV NSs and RVFV NSs to promote PKR downregulation, we infected 293 cells with either wild-type MP-12 or rMP12-TOSNSs at an MOI of 3 and harvested cells at 0, 2, 4, 6, 8, 10, 12, and 24 hpi for analysis by Western blotting with anti-PKR, anti-TOSV NSs, and anti-RVFV antibodies (Fig. 3B). Both viruses downregulated PKR with similar kinetics, and just as was observed for wild-type TOSV infection, the majority of PKR was degraded between 8 and 10 hpi. Also, similar to the expression of TOSV NSs in the context of the authentic virus, both TOSV and RVFV NSs proteins could be detected as early as 4 hpi when expressed in the MP-12 background. In contrast, when cells were mock infected or infected with rMP12-C13type, no change in the expression of PKR could be detected, suggesting that the NSs protein, and not another product of viral replication, is responsible for mediating the downregulation of PKR.
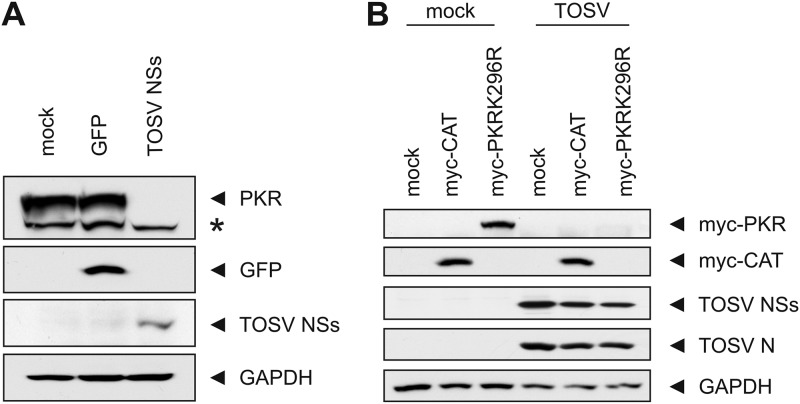
To determine whether the TOSV NSs protein alone was sufficient to downregulate PKR, we transfected 293 cells with in vitro-synthesized RNA encoding NSs (Fig. 4A). To exclude the possibility that the observed PKR downregulation was an unspecific effect of transfected RNA, we also transfected in vitro-synthesized RNA for GFP as a negative control. Whole-cell lysates from cells collected at 20 h after transfection were analyzed by Western blotting with anti-PKR, anti-GFP, anti-TOSV, and anti-GAPDH antibodies. We observed a downregulation of PKR only in cells transfected with RNA encoding TOSV NSs, but not GFP, although both proteins were expressed. In summary, these results indicate that TOSV is able to downregulate PKR as efficiently as RVFV and that the TOSV NSs protein is able to promote PKR downregulation regardless of whether it is expressed by authentic TOSV or in the RVFV MP-12 background or expressed on its own.
Fig 4.
PKR downregulation by TOSV NSs. (A) TOSV NSs mediates PKR degradation. 293 cells were mock transfected or transfected with in vitro-synthesized RNA for GFP (control) or TOSV NSs and harvested at 20 h posttransfection. Whole-cell lysates were analyzed by Western blotting with anti-PKR, anti-GFP, anti-TOSV, or anti-GAPDH antibodies (from top to bottom), respectively. The asterisk denotes a nonspecific band. (B) Activation of PKR is not required for downregulation. 293 cells were mock infected or infected with TOSV at an MOI of 3 and transfected with in vitro-synthesized RNA encoding myc-PKRK296R, a kinase-inactive form of PKR, or myc-CAT immediately after virus adsorption. Whole-cell lysates were collected at 16 hpi and analyzed by Western blotting with anti-myc, anti-TOSV NSs, anti-TOSV, or anti-GAPDH antibodies (from top to bottom), respectively.
Phosphorylation of PKR is not required for PKR degradation by TOSV NSs.
We next determined whether phosphorylation of PKR was required for its TOSV NSs-mediated downregulation. To do this, we infected cells with TOSV at an MOI of 3 and transfected them with in vitro-synthesized RNA encoding a myc-tagged, kinase-inactivated form of PKR (myc-PKRK296R) (32) immediately after virus adsorption. As a control to show that TOSV replication did not interfere with RNA transfection or translation from the transfected constructs, we also transfected cells with myc-CAT. Figure 4B shows that the kinase-inactive K296R PKR is still downregulated by TOSV, whereas myc-CAT expression remains unaffected by viral replication. This result indicates that phosphorylation of PKR is not a prerequisite for its downregulation by TOSV NSs.
TOSV promotes the posttranslational degradation of PKR.
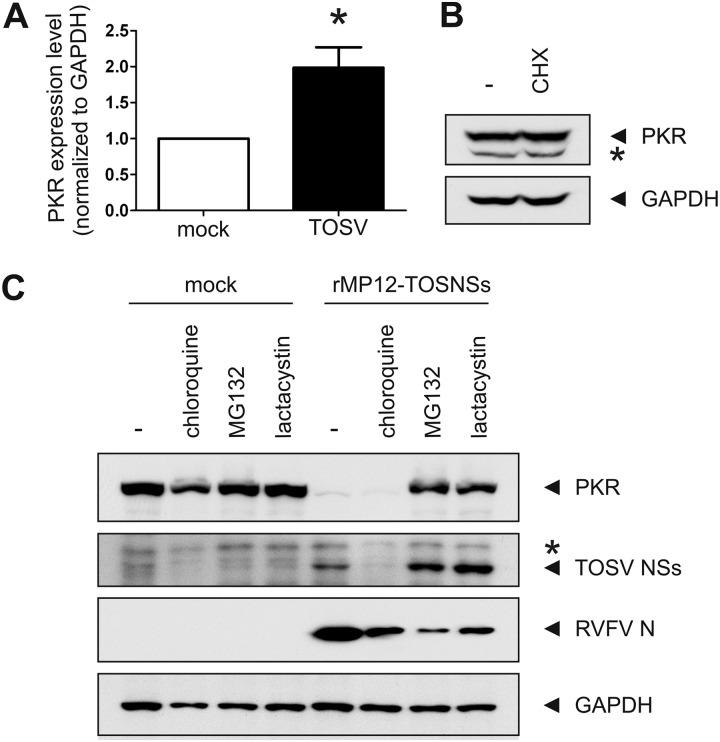
Having established that the TOSV NSs protein was able to promote the downregulation of PKR, we next set out to determine whether this was caused at the level of mRNA expression, protein synthesis, or posttranslational degradation. To determine whether NSs was able to modulate the expression of PKR mRNA, we used quantitative real-time PCR to determine the relative expression of PKR mRNA compared to that of GAPDH mRNA in cells which had been infected with TOSV for 16 h (Fig. 5A). Interestingly, the expression of PKR mRNA was even increased (P < 0.05) in comparison to mock-infected cells, although no PKR protein remained this late during TOSV infection (Fig. 3A). Similar results were also obtained for cells collected at 8 hpi (data not shown). This indicates that TOSV NSs does not regulate PKR expression at the level of mRNA.
Fig 5.
The proteasome is involved in the TOSV NSs-mediated downregulation of PKR. (A) PKR downregulation is not mediated on mRNA level. 293 cells were mock infected or infected with TOSV at an MOI of 3. Total RNA was extracted after harvesting at 16 hpi, and the relative expression of PKR mRNA was determined by quantitative real-time PCR. An asterisk (*) indicates statistical significance (paired t test, P < 0.05). (B) PKR is degraded through a posttranslational mechanism. 293 cells were mock treated or treated with 100 μg of CHX/ml for 8 h to inhibit translation. Whole-cell lysates were analyzed by Western blotting with anti-PKR (top) or anti-GAPDH (bottom) antibodies, respectively. The asterisk denotes a nonspecific band. (C) PKR is stabilized under proteasome impairment. 293 cells were mock infected or infected with rMP12-TOSNSs at an MOI of 3. At 2 hpi, the cells were treated with 100 μM chloroquine, 5 μM MG132, or 50 mM lactacystin where indicated. Whole-cell lysates were collected at 20 hpi and analyzed by Western blotting with anti-PKR, anti-TOSV, anti-RVFV, or anti-GAPDH (from top to bottom) antibodies, respectively. The asterisk denotes a nonspecific band.
We next used pharmacological inhibition of protein synthesis to determine whether NSs affected PKR expression at the level of translation. We treated uninfected 293 cells with cycloheximide (CHX) for 8 h before determining the amount of remaining PKR by Western blotting (Fig. 5B). We reasoned that if NSs was able to inhibit PKR translation, pharmacological inhibition of protein synthesis for 8 h—a time point during TOSV infection at which only marginal amounts of PKR remain—should also result in decreased levels of PKR. However, treatment of 293 cells with CHX had no effect on PKR levels, indicating that PKR turnover in these cells is rather slow and that inhibition of translation for 8 h, whether pharmacological or virus-mediated, has no effect on cellular PKR levels. Thus, TOSV NSs most likely downregulates PKR at the posttranslational level rather than at the transcriptional or translational level.
TOSV NSs promotes the proteasomal degradation of PKR.
In order to identify the protease responsible for the NSs-mediated downregulation of PKR, we infected 293 cells with rMP12-TOSNSs at an MOI of 3. At 2 hpi, the cells were treated with either lysosomal or proteasomal inhibitors and harvested at 20 hpi. Whole-cell lysates were then analyzed by Western blotting with anti-PKR, anti-TOSV (to detect the expression of TOSV NSs), anti-RVFV (to detect the expression of RVFV N), and anti-GAPDH antibodies (Fig. 5C). Treatment of cells with the lysosomal inhibitor chloroquine had no effect on the NSs-mediated degradation of PKR, even though it led to a reduction in the expression of TOSV NSs. This effect has also been described for MP-12 NSs (13). Treatment of cells with the proteasomal inhibitors MG132 and lactacystin, on the other hand, was able to stabilize PKR expression during infection with rMP12-TOSNSs. Interestingly, inhibition of the proteasome increased the levels of TOSV NSs in infected cells, suggesting that NSs itself is a substrate of the proteasome. In contrast, both the N and the NSs proteins of RVFV are equally affected by proteasomal inhibition (13). We also used wild-type TOSV in a similar experiment, and yet wild-type TOSV did not replicate in the presence of any of the three inhibitors used, and when these inhibitors were added at later times during infection, the PKR levels had already been reduced (data not shown). These data suggest that the proteasome is involved in the TOSV NSs-mediated degradation of PKR as well the turnover of NSs itself.
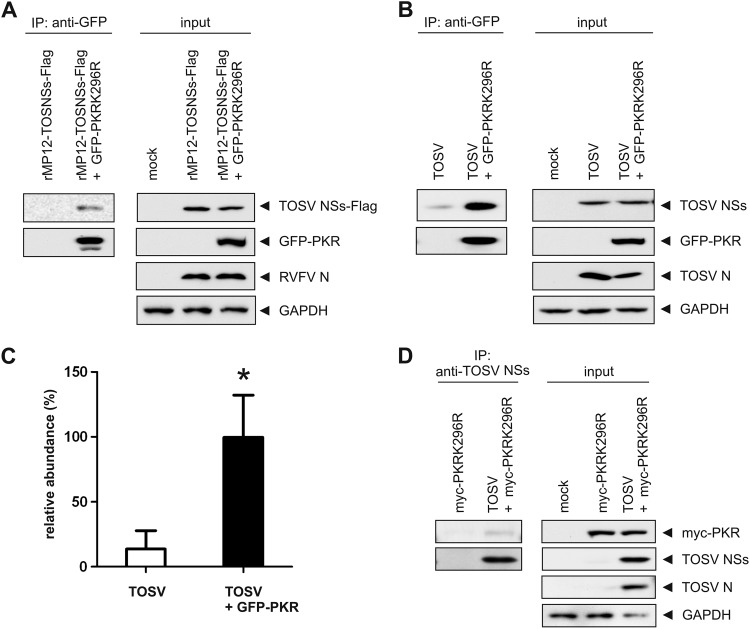
TOSV NSs protein interacts with PKR in infected cells.
TOSV NSs could potentially target PKR directly to the proteasome or promote its degradation through subverting a thus-far-unidentified host factor responsible for mediating PKR degradation. We therefore investigated whether NSs had the ability to interact with PKR in infected cells, which would suggest a direct targeting mechanism. To this aim, we transiently transfected cells with a kinase-inactive, GFP-tagged PKR (GFP-PKRK296R) and subsequently infected them with a recombinant MP-12 encoding TOSV NSs with a Flag tag at the C terminus (rMP12-TOSNSs-Flag) at an MOI of 3 (Fig. 6A). We used kinase-inactive PKR because the overexpression of wild-type PKR inhibited viral replication (data not shown). TOSV NSs tagged with Flag was used in this experiment to facilitate detection of TOSV NSs, and we confirmed that rMP12-TOSNSs-Flag was able to promote PKR degradation (data not shown). Lysates of infected cells were subjected to immunoprecipitation using anti-GFP magnetic beads and subsequent analysis by Western blotting with anti-FLAG, anti-GFP, and anti-RVFV antibodies. As shown in Fig. 6A, Flag-tagged TOSV NSs could be coprecipitated with the anti-GFP antibody when cells had been transfected with GFP-PKRK296R, but not in cells only infected with rMP12-TOSNSs-Flag. To determine whether authentic TOSV NSs could also be coprecipitated with PKR, we transfected cells with GFP-PKRK296R, and then infected them with TOSV at an MOI of 3. GFP-PKR was precipitated as described above, and the precipitates were analyzed by Western blotting with anti-TOSV NSs, anti-GFP, and anti-TOSV antibodies (Fig. 6B). As was observed for the Flag-tagged TOSV NSs, authentic TOSV NSs could be coprecipitated with the anti-GFP antibody when cells had been transfected with GFP-PKRK296R. Since we repeatedly observed a background level of coprecipitated TOSV NSs with anti-GFP antibody when detected with the anti-TOSV antibody, we compared the relative abundance of TOSV NSs between cells mock-transfected and those transfected with myc-PKRK296R. This revealed that significantly more TOSV NSs was coprecipitated when cells were expressing GFP-PKRK296R (Fig. 6C), suggesting that TOSV NSs is able to interact with PKR-GFPK296R. To determine whether PKR could reciprocally be coprecipitated with TOSV NSs, we transiently transfected cells with a kinase-inactive, myc-tagged PKR (myc-PKRK296R) and subsequently infected them with TOSV at an MOI of 3. Lysates of infected cells were subjected to immunoprecipitation using anti-TOSV NSs antibodies and protein G-magnetic beads and subsequent analysis by Western blotting with anti-myc, anti-TOSV NSs, and anti-TOSV antibodies. As shown in Fig. 6D, the anti-TOSV NSs antibody was able to coprecipitate myc-PKRK296R from TOSV-infected cells but not from mock-infected cells. In conclusion, these results demonstrate that TOSV NSs is able to interact with kinase-inactive PKR in infected cells.
Fig 6.
TOSV NSs interacts with PKR in infected cells. (A) 293 cells were mock transfected or transfected with GFP-PKRK296R and subsequently infected with rMP12-TOSNSs-Flag. The cells were then harvested and subjected to anti-GFP immunoprecipitation. The precipitates (left panel) and 10% of the input lysates (right panel) were analyzed by Western blotting with anti-FLAG, anti-GFP, anti-RVFV, or anti-GAPDH (from top to bottom) antibodies, respectively. (B) 293 cells were mock transfected or transfected with GFP-PKRK296R and subsequently infected with TOSV. The cells were then harvested and subjected to immunoprecipitation with anti-GFP antibody. The precipitates (left panel) and 10% of the input lysates (right panel) were analyzed by Western blotting with anti-TOSV NSs, anti-GFP, anti-TOSV, or anti-GAPDH (from top to bottom) antibodies, respectively. (C) Quantification of the relative abundance of TOSV NSs after immunoprecipitation with anti-GFP antibody shown in panel B. Shown are the means and SD of three independent experiments. The mean value of TOSV + GFP-PKR was set to 100%. The asterisk (*) indicates statistical significance (paired Student t test, P < 0.05). (D) 293 cells were mock transfected or transfected with myc-PKRK296R and subsequently infected with TOSV. The cells were then harvested and subjected to immunoprecipitation with anti-TOSV NSs antibody. The precipitates (left panel) and 10% of the input lysates (right panel) were analyzed by Western blotting with anti-myc, anti-TOSV NSs, anti-TOSV, or anti-GAPDH (from top to bottom) antibodies, respectively.
DISCUSSION
Members of the genus Phlebovirus have a wide spectrum of pathogenicity. Viruses that are able to productively infect humans can cause symptoms ranging from a mild, self-limiting illness of short duration (such as SFSV or PTV) to a potentially fatal disease (such as RVFV) with fulminant hepatitis and encephalitic manifestations (2). Interestingly, the NSs protein, which has been identified as the major virulence factor in these viruses, is the least conserved among proteins encoded by these viruses, with sequence similarities ranging from only 30% to below 17%. In particular, the NSs proteins of RVFV and TOSV share only 48 positions (18.1%) and differ in size by 51 amino acids, with TOSV NSs being almost 20% longer than RVFV NSs (3). Conversely, the N proteins of different phleboviruses share up to 54% similarity and the RVFV N protein in particular is only 8 amino acids (3.3%) longer than that of TOSV (3). Further studies will be required to understand the differences among the phlebovirus NSs proteins in detail, including the subcellular localization of TOSV, PTV, SFSV, and FRIV NSs, the accumulation kinetics of PTV, SFSV, or FRIV NSs, and other functions of these proteins.
The NSs protein of RVFV has multiple functions: (i) general transcription suppression by sequestering TFIIH p44 (33) and by promoting the degradation of TFIIH p62 (13), (ii) IFN-β promoter suppression by specific binding to SAP30 (11), and (iii) PKR degradation (14, 15). In a previous study, TOSV NSs has been shown to act as an IFN-β antagonist when expressed from a transfected plasmid or in the context of a recombinant RVFV, even though replication of authentic TOSV still induced IFN-β (19). In the present study, we discovered that TOSV NSs promotes PKR degradation, while host transcription is not suppressed by TOSV NSs. Thus, TOSV NSs encodes two distinct functions: (i) to limit IFN-β upregulation (19) and (ii) to promote PKR degradation. Considering that TOSV NSs does not inhibit host general transcription, cells infected with TOSV may induce cytokines or other cell mediators in response to TOSV replication, which are different from those produced in cells infected with RVFV. This induction may be important for the pathogenesis of TOSV but, unfortunately, there are no rodent models available for pathological studies except for a mouse intracerebral inoculation model with a neuro-adapted strain of TOSV (34). The recombinant MP-12 or wild-type ZH501 encoding TOSV NSs may be useful to understand the role of TOSV NSs in the neuroinvasiveness and increased virulence of TOSV in comparison to other phlebotomus fevers.
We found that the abundance of TOSV NSs was increased in cells treated with proteasomal inhibitors. This is in contrast to RVFV N and NSs, the expression of which is negatively affected by the treatment of cells with proteasome inhibitors (13), an effect which is most likely mediated by activation of the unfolded protein response due to the accumulation of misfolded proteins (35), and the resulting shutoff cellular translation (36). It is likely that TOSV NSs translation is also negatively affected by treatment with proteasome inhibitors, but that at the same time the protein is stabilized by inhibition of the proteasome. If TOSV NSs has a rapid turnover time, the stabilization by proteasomal inhibitors would outweigh the effect of reduced translation, and ultimately result in higher abundance of NSs under these conditions, as we have observed here. The finding that both PKR and TOSV NSs are stabilized under proteasome inhibition suggests that TOSV NSs might be degraded along with its substrate and thus utilize a mechanism that is distinct from that of RVFV NSs-mediated PKR degradation.
We demonstrated that TOSV NSs could promote the downregulation of a kinase-inactive PKR, suggesting that phosphorylation of PKR is not required for its NSs-mediated downregulation. Furthermore, we demonstrated that TOSV NSs interacted with kinase-inactive GFP-PKRK296R in infected cells, suggesting that TOSV NSs forms a complex with PKR to target it for proteasomal degradation. Further studies will be required to determine whether NSs promotes PKR polyubiquitination, and to identify the host proteins interacting with the NSs-PKR complex, which will help elucidate the mechanism of PKR degradation by the NSs proteins of different phleboviruses.
In summary, we found that TOSV NSs can promote the degradation of PKR, while not affecting host general transcription. This novel finding will be important to understand the unique pathogenesis of TOSV infection in humans.
ACKNOWLEDGMENTS
We thank R. B. Tesh for providing TOSV (ISS.Phl.3), PTV (D-4021A), FRIV (VP-161A), SFSV (Sabin), and antisera against RVFV, TOSV, PTV, FRIV, and SFSV.
This study was supported by 5 U54 AI057156 through the Western Regional Center of Excellence, NIH grant R01 AI08764301, and funding from the Sealy Center for Vaccine Development at UTMB. B.K. was supported by the James W. McLaughlin Fellowship Fund at the University of Texas Medical Branch.
Footnotes
Published ahead of print 16 January 2013
REFERENCES
- 1. Xu F, Liu D, Nunes M, Tesh RB, Xiao SY. 2007. Antigenic and genetic relationships among Rift Valley fever virus and other selected members of the genus Phlebovirus (Bunyaviridae). Am. J. Trop. Med. Hyg. 76:1194–1200 [PubMed] [Google Scholar]
- 2. Schmaljohn C, Nichol S. 2007. Bunyaviruses, p 1741–1789 In Knipe D, Howley P, Griffin D, Lamb R, Martin A. (ed), Fields virology, 5th ed Lippincott/The Williams & Wilkins Co, Philadelphia, PA [Google Scholar]
- 3. Giorgi C, Accardi L, Nicoletti L, Gro MC, Takehara K, Hilditch C, Morikawa S, Bishop DH. 1991. Sequences and coding strategies of the S RNAs of Toscana and Rift Valley fever viruses compared to those of Punta Toro, Sicilian sandfly fever, and Uukuniemi viruses. Virology 180:738–753 [DOI] [PubMed] [Google Scholar]
- 4. Charrel RN, Gallian P, Navarro-Mari JM, Nicoletti L, Papa A, Sanchez-Seco MP, Tenorio A, de Lamballerie X. 2005. Emergence of Toscana virus in Europe. Emerg. Infect. Dis. 11:1657–1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dionisio D, Valassina M, Ciufolini MG, Vivarelli A, Esperti F, Cusi MG, Marchi A, Mazzoli F, Lupi C. 2001. Encephalitis without meningitis due to sandfly fever virus serotype Toscana. Clin. Infect. Dis. 32:1241–1243 [DOI] [PubMed] [Google Scholar]
- 6. Nicoletti L, Verani P, Caciolli S, Ciufolini MG, Renzi A, Bartolozzi D, Paci P, Leoncini F, Padovani P, Traini E. 1991. Central nervous system involvement during infection by Phlebovirus Toscana of residents in natural foci in central Italy (1977–1988). Am. J. Trop. Med. Hyg. 45:429–434 [DOI] [PubMed] [Google Scholar]
- 7. Valassina M, Cuppone AM, Bianchi S, Santini L, Cusi MG. 1998. Evidence of Toscana virus variants circulating in Tuscany, Italy, during the summers of 1995 to 1997. J. Clin. Microbiol. 36:2103–2104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tesh RB. 1988. The genus Phlebovirus and its vectors. Annu. Rev. Entomol. 33:169–181 [DOI] [PubMed] [Google Scholar]
- 9. Boshra H, Lorenzo G, Busquets N, Brun A. 2011. Rift Valley fever: recent insights into pathogenesis and prevention. J. Virol. 85:6098–6105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cusi MG, Savellini GG, Zanelli G. 2010. Toscana virus epidemiology: from Italy to beyond. Open Virol. J. 4:22–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Le May N, Mansuroglu Z, Leger P, Josse T, Blot G, Billecocq A, Flick R, Jacob Y, Bonnefoy E, Bouloy M. 2008. A SAP30 complex inhibits IFN-beta expression in Rift Valley fever virus infected cells. PLoS Pathog. 4:e13 doi:10.1371/journal.ppat.0040013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Le May N, Dubaele S, Proietti De Santis L, Billecocq A, Bouloy M, Egly JM. 2004. TFIIH transcription factor, a target for the Rift Valley hemorrhagic fever virus. Cell 116:541–550 [DOI] [PubMed] [Google Scholar]
- 13. Kalveram B, Lihoradova O, Ikegami T. 2011. NSs protein of Rift Valley fever virus promotes posttranslational downregulation of the TFIIH subunit p62. J. Virol. 85:6234–6243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Habjan M, Pichlmair A, Elliott RM, Overby AK, Glatter T, Gstaiger M, Superti-Furga G, Unger H, Weber F. 2009. NSs protein of Rift Valley fever virus induces the specific degradation of the double-stranded RNA-dependent protein kinase. J. Virol. 83:4365–4375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ikegami T, Narayanan K, Won S, Kamitani W, Peters CJ, Makino S. 2009. Rift Valley fever virus NSs protein promotes posttranscriptional downregulation of protein kinase PKR and inhibits eIF2α phosphorylation. PLoS Pathog. 5:e1000287 doi:10.1371/journal.ppat.1000287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mansuroglu Z, Josse T, Gilleron J, Billecocq A, Leger P, Bouloy M, Bonnefoy E. 2010. Nonstructural NSs protein of Rift Valley fever virus interacts with pericentromeric DNA sequences of the host cell, inducing chromosome cohesion and segregation defects. J. Virol. 84:928–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Baer A, Austin D, Narayanan A, Popova T, Kainulainen M, Bailey C, Kashanchi F, Weber F, Kehn-Hall K. 2012. Induction of DNA damage signaling upon Rift Valley fever virus infection results in cell cycle arrest and increased viral replication. J. Biol. Chem. 287:7399–7410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Perrone LA, Narayanan K, Worthy M, Peters CJ. 2007. The S segment of Punta Toro virus (Bunyaviridae, Phlebovirus) is a major determinant of lethality in the Syrian hamster and codes for a type I interferon antagonist. J. Virol. 81:884–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gori Savellini G, Weber F, Terrosi C, Habjan M, Martorelli B, Cusi MG. 2011. Toscana virus induces interferon although its NSs protein reveals antagonistic activity. J. Gen. Virol. 92:71–79 [DOI] [PubMed] [Google Scholar]
- 20. Garcia MA, Gil J, Ventoso I, Guerra S, Domingo E, Rivas C, Esteban M. 2006. Impact of protein kinase PKR in cell biology: from antiviral to antiproliferative action. Microbiol. Mol. Biol. Rev. 70:1032–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ito N, Takayama-Ito M, Yamada K, Hosokawa J, Sugiyama M, Minamoto N. 2003. Improved recovery of rabies virus from cloned cDNA using a vaccinia virus-free reverse genetics system. Microbiol. Immunol. 47:613–617 [DOI] [PubMed] [Google Scholar]
- 22. Caplen H, Peters CJ, Bishop DH. 1985. Mutagen-directed attenuation of Rift Valley fever virus as a method for vaccine development. J. Gen. Virol. 66:2271–2277 [DOI] [PubMed] [Google Scholar]
- 23. Ikegami T, Won S, Peters CJ, Makino S. 2006. Rescue of infectious Rift Valley fever virus entirely from cDNA, analysis of virus lacking the NSs gene, and expression of a foreign gene. J. Virol. 80:2933–2940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kalveram B, Lihoradova O, Indran SV, Ikegami T. 2011. Using reverse genetics to manipulate the NSs gene of the Rift Valley fever virus MP-12 strain to improve vaccine safety and efficacy. J. Vis. Exp. 2011:e3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zamoto-Niikura A, Terasaki K, Ikegami T, Peters CJ, Makino S. 2009. Rift Valley fever virus L protein forms a biologically active oligomer. J. Virol. 83:12779–12789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Billecocq A, Gauliard N, Le May N, Elliott RM, Flick R, Bouloy M. 2008. RNA polymerase I-mediated expression of viral RNA for the rescue of infectious virulent and avirulent Rift Valley fever viruses. Virology 378:377–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Vialat P, Muller R, Vu TH, Prehaud C, Bouloy M. 1997. Mapping of the mutations present in the genome of the Rift Valley fever virus attenuated MP12 strain and their putative role in attenuation. Virus Res. 52:43–50 [DOI] [PubMed] [Google Scholar]
- 28. Sadler AJ, Williams BR. 2007. Structure and function of the protein kinase R. Curr. Top. Microbiol. Immunol. 316:253–292 [DOI] [PubMed] [Google Scholar]
- 29. Diaz MO, Ziemin S, Le Beau MM, Pitha P, Smith SD, Chilcote RR, Rowley JD. 1988. Homozygous deletion of the alpha- and beta 1-interferon genes in human leukemia and derived cell lines. Proc. Natl. Acad. Sci. U. S. A. 85:5259–5263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mosca JD, Pitha PM. 1986. Transcriptional and posttranscriptional regulation of exogenous human beta interferon gene in simian cells defective in interferon synthesis. Mol. Cell. Biol. 6:2279–2283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jao CY, Salic A. 2008. Exploring RNA transcription and turnover in vivo by using click chemistry. Proc. Natl. Acad. Sci. U. S. A. 105:15779–15784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cosentino GP, Venkatesan S, Serluca FC, Green SR, Mathews MB, Sonenberg N. 1995. Double-stranded-RNA-dependent protein kinase and TAR RNA-binding protein form homo- and heterodimers in vivo. Proc. Natl. Acad. Sci. U. S. A. 92:9445–9449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Billecocq A, Spiegel M, Vialat P, Kohl A, Weber F, Bouloy M, Haller O. 2004. NSs protein of Rift Valley fever virus blocks interferon production by inhibiting host gene transcription. J. Virol. 78:9798–9806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cusi MG, Gori Savellini G, Terrosi C, Di Genova G, Valassina M, Valentini M, Bartolommei S, Miracco C. 2005. Development of a mouse model for the study of Toscana virus pathogenesis. Virology 333:66–73 [DOI] [PubMed] [Google Scholar]
- 35. Obeng EA, Carlson LM, Gutman DM, Harrington WJ, Jr, Lee KP, Boise LH. 2006. Proteasome inhibitors induce a terminal unfolded protein response in multiple myeloma cells. Blood 107:4907–4916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhao L, Ackerman SL. 2006. Endoplasmic reticulum stress in health and disease. Curr. Opin. Cell Biol. 18:444–452 [DOI] [PubMed] [Google Scholar]