Abstract
Coxsackievirus A9 (CVA9) is an important pathogen of the Picornaviridae family. It utilizes cellular receptors from the integrin αv family for binding to its host cells prior to entry and genome release. Among the integrins tested, it has the highest affinity for αvβ6, which recognizes the arginine-glycine-aspartic acid (RGD) loop present on the C terminus of viral capsid protein, VP1. As the atomic model of CVA9 lacks the RGD loop, we used surface plasmon resonance, electron cryo-microscopy, and image reconstruction to characterize the capsid-integrin interactions and the conformational changes on genome release. We show that the integrin binds to the capsid with nanomolar affinity and that the binding of integrin to the virion does not induce uncoating, thereby implying that further steps are required for release of the genome. Electron cryo-tomography and single-particle image reconstruction revealed variation in the number and conformation of the integrins bound to the capsid, with the integrin footprint mapping close to the predicted site for the exposed RGD loop on VP1. Comparison of empty and RNA-filled capsid reconstructions showed that the capsid undergoes conformational changes when the genome is released, so that the RNA-capsid interactions in the N termini of VP1 and VP4 are lost, VP4 is removed, and the capsid becomes more porous, as has been reported for poliovirus 1, human rhinovirus 2, enterovirus 71, and coxsackievirus A7. These results are important for understanding the structural basis of integrin binding to CVA9 and the molecular events leading to CVA9 cell entry and uncoating.
INTRODUCTION
Picornaviruses that belong to the Human enterovirus B species (HEV-B) in the family Picornaviridae are the major cause of aseptic meningitis (1, 2). HEV-B types, including coxsackievirus A9 (CVA9), also exhibit a wide range of other clinical manifestations of acute disease, including respiratory infections, myocarditis, encephalitis, paralysis, rash, and severe generalized infections in newborns (3–7). Moreover, there is evidence that suggests that HEV-B, specifically, CVA9, CVB1, CVB3, and CVB5, may be involved in the pathogenesis of childhood diabetes, the rate of incidence of which is increasing in Western countries (8–11).
The CVA9 particle is about 28 nm in diameter and consists of a nonenveloped capsid with icosahedral symmetry, surrounding a positive-sense RNA genome of 7,452 nucleotides (12, 13). The CVA9 T=1 (pseudo-T=3) capsid is made up of 60 copies each of the four viral capsid proteins (VP1 to VP4) as revealed by its atomic model (12). Each of VP1, VP2, and VP3 possesses an eight-stranded β-barrel structure. VP1 mainly forms the vertices of the capsid, with VP2 and VP3 alternating on the 3-fold and 2-fold axes of symmetry (12). These are common characteristics that are shared by most picornaviruses (12, 14). Depressions of about 15 Å form the canyon region and are also seen around the 2-fold axes (12). CVA9 interacts with cell surface αv integrins during the early stages of infection via an arginine-glycine-aspartic acid (RGD) motif in VP1 (13, 15–17). Sequence analysis of clinical CVA9 isolates spanning almost 50 years has indicated that the RGD motif is conserved, suggesting that the integrin interaction(s) is important in viral infection (18), even though RGD-less CVA9 has been grown in some cell lines (19). The C-terminal 15 amino acids of VP1 containing the RGD motif could not be identified in the X-ray structure of CVA9 (12).
Integrins are members of a family of cellular membrane receptors found as heterodimers of an α- and a β-chain. The heterodimer body can be divided into a head piece and a tail piece, with three known conformations of extended open head piece, extended closed head piece, and bent head piece (20). For leukocyte integrins, the bent physiological state (low affinity) can bind a Fab that then induces the extended, activated states with high affinity (21). A cyclic RGD peptide has been shown to bind the integrin αvβ3 head piece in the β-propeller region of the αv chain through arginine and in the I-like region of the β3 chain through the aspartic acid (22). Adenovirus has been modeled with αvβ5 bound in an extended conformation (23). So far, there has been no structural description of the interaction between CVA9 and any of the αv integrin heterodimers. Biochemically, it has been shown that CVA9 binds to both integrin αvβ3 and integrin αvβ6 in vitro (16, 17), with preferential binding to αvβ6.
The integrin αVβ6 is thought to be a high-affinity binding receptor for the virus (15, 24), rather than signaling directly the endocytosis of the virus, because the virus has not been observed to colocalize with internalized integrin (15, 25). Furthermore, inhibition of the downstream signaling pathways of αv integrin does not appear to affect virus proliferation (15). These data indicate that integrin αvβ6 acts in the early steps of the virus life cycle to initiate binding of the virus particles to the cell surface; entry requires other host cell factors such as β2-microglobulin and GRP78 (15, 26). It is, however, unclear at what stage the viral RNA is released into the cytosol, and whether interaction with the receptor (αvβ6) plays a role in this process.
There are 60 potential receptor binding sites for the integrin on the picornaviral capsid surface (one per VP1 molecule), but previous studies of other virus-integrin complexes by electron cryo-microscopy (cryo-EM) and icosahedral reconstruction have indicated that only some of the sites are occupied, as the intensity for the integrin is much weaker than that for the rest of the capsid (23, 27, 28). One way of judging the occupancy at equivalent sites on the surface of a highly symmetric object is to carry out tomographic reconstructions of individual viruses (28); another is to calculate an asymmetric reconstruction (29, 30). The occupancy may be affected by steric hindrance or the conformation of the ligand, which may be revealed by studying the binding kinetics in more detail. In surface plasmon resonance (SPR) measurements, one of the binding partners is immobilized on the chip while the other is passed over it in an excess to establish the binding kinetics. This approach has been successful for binding studies of adenovirus with integrin and rhinovirus with ICAM-1 (27, 31).
CVA9 is one of the few picornaviruses besides foot-and-mouth disease virus (32), echovirus 1 (33), echovirus 9 (34), and human parechovirus 1 (35) that is known to use integrins as a cell surface receptor. The rather unique involvement of integrin αVβ6 in CVA9 binding motivated us to analyze the interaction in higher detail at the structural level. Using cryo-EM, tomography, and three-dimensional image reconstruction by the use of icosahedral symmetry and asymmetric reconstruction, and fitting of the X-ray model, we identified the footprints of integrin on the capsid surface and in regions where the capsid proteins interact with RNA. In addition, we analyzed the movements of the capsid proteins VP1, VP2, and VP3 upon release of the genome from the virion by comparison of empty and RNA-filled capsids.
MATERIALS AND METHODS
Purification of coxsackievirus A9 and integrin αVβ6.
Methods for production and purification of coxsackievirus A9 and the ectodomain of integrin αvβ6 have been described earlier (12, 28).
Cryo-EM and cryo-ET.
CVA9 capsids were mixed with integrin αvβ6 at 1:265 and 1:100 molar ratios of capsid to integrin for 1 h at room temperature prior to cryosample preparation on glow-discharged Quantifoil R2/2 (Quantifoil Micro Tools GmbH, Germany) and C-Flat 224 (Electron Microscopy Sciences) grids as described previously (36). Control samples were prepared similarly. Colloidal gold particles (10-nm diameter) were added to the samples as fiducial markers for electron tomography. The micrographs for icosahedral and asymmetric reconstruction were imaged using an electron dose of ∼15 to 20 e− Å−2 at a magnification of ×62,000 on film and scanned to result in a sampling size of 0.113 nm/pixel as described previously (28). For electron cryo-tomography (cryo-ET), six tilt series (±60° at 3° increments) at an underfocus of ∼8 μm were collected with Serial EM software on a Tecnai F20 microscope maintained at liquid nitrogen temperature using a Gatan Ultrascan 4000 charge-coupled device (37). A nominal magnification of ×39,400 was used for data collection, giving a sampling size of 0.38 nm/pixel.
Image processing for icosahedral reconstruction and tomographic reconstruction.
Micrographs were discarded if they had noticeable drift or astigmatism, or contained crystalline ice. The defocus level of each micrograph was determined using CTFFIND3 (38). Particles were automatically picked using ETHAN (39) and then manually screened, separated into filled and empty particle data sets (micrographs included both particle types), and windowed in EMAN (40). Starting models for both data sets were generated using a random model computation procedure (41, 42) operating on 150 images selected from the furthest-from-focus micrographs and resulted in initial reconstructions at ∼30 Å. These reconstructions then served as starting models for full orientation and origin determination of the entire data set using AUTO3DEM (41). The resolution of the reconstructions was estimated by Fourier shell correlation analysis with a threshold criterion of 0.5 (43). The statistics for the reconstructions are collated in Table 1. The radial program from the BSOFT (44) package was used to calculate the radial profiles.
Table 1.
Statistics for the icosahedrally symmetric reconstructions
| Parameter | Value(s) |
|||
|---|---|---|---|---|
| Filled CVA9 reconstruction | Empty CVA9 reconstruction | Filled CVA9 integrin αvβ6 | Empty CVA9 integrin αvβ6 | |
| No. of micrographs | 56 | 56 | 147 | 147 |
| No. of particles used in the reconstruction | 488 | 728 | 1,597 | 1,200 |
| Defocus range (μm) | 0.89–2.32 | 0.89–2.32 | 0.83–4.12 | 0.83–4.12 |
| Sampling (nm/pixel) | 0.113 | 0.113 | 0.226 | 0.226 |
| Resolution (Å) | 10.3 | 9.9 | 9.0 | 9.5 |
Tomographic data were binned by a factor of 2. Tomograms were reconstructed using IMOD software (45), truncating the data to the first zero of the contrast transfer function to remove information beyond 4.4 nm. A nonlinear anisotropic diffusion filter was applied for visualization.
Asymmetric reconstruction of filled CVA9-integrin αVβ6 complex.
The asymmetric reconstruction was carried out as described earlier for bacteriophage Sf6 (30). First, a model of the integrin ligand in the activated state was created, combining the coordinates for the integrin αIIbβ3 complete head piece (Protein Data Bank [PDB] 3FCS) (46) for the inactive state and the partial structure in the open state (PDB 3FCU) (46) by simply extending the open-state-structure α-chain (for which there is only the β-propeller structure) from the inactive state by aligning these two, while keeping the β-chain at the open-state-structure angle to the α-chain. The α-chain was assumed to have the “activated” extended structure, which was generated manually by removing the kink from the inactive-state structure by rotation and translation of the α-chain domains with PyMol (The PyMOL Molecular Graphics System, Version 1.5.0.4; Schrödinger, LLC). For further processing, we used only the β-chain and generated a reference model by combining the icosahedrally symmetric reconstruction of the filled capsid with one molecule of the β-chain attached to one of the 60 equivalent positions indicated by the icosahedrally symmetric reconstruction of the filled capsid complexed with integrin. This model was used as the starting map in AUTO3DEM for a data set of close-to-focus (0.83- to 1.54-μm) particles. The data were binned by a factor of 2, where the integrin density is more readily visible. The icosahedral origins and orientations already determined for CVA9 filled capsid-integrin αvβ6 images were used as the starting point, where the few positions with integrins bound are not all aligned. In the AUTO3DEM file, the symmetry code was changed from 532 (icosahedral) to 1 (asymmetric) and the ticos_equivalent option was activated (= 1) to search for the best of the 60 possible equivalent positions. AUTO3DEM was then allowed to run in refine mode for a single iteration with a fixed center. The expected result was that one of the 60 possible positions where there was an integrin bound for each particle would align to the reference. The other occupied positions would either show some preference for occupancy (high signal seen on another capsid position) or still be randomly attached (low signal seen on other capsid positions). Control iterations were run with both integrin chains present in the correct position or only the β-chain attached to the 5-fold vertices on the capsid (an incorrect position), to investigate model bias.
Fitting of CVA9 capsid proteins.
The X-ray structure of CVA9 (PDB 1d4m) (12) was placed in both the filled CVA9-integrin αvβ6 density map and the empty CVA9-integrin αvβ6 map. Additional rigid and flexible body fitting was carried out for the empty CVA9-integrin αvβ6 density map. The capsid map containing the X-ray structure of CVA9 was zoned to a single asymmetric unit using the Zone feature in Chimera (47) with a zoning radius of 12 Å, followed by flexible fitting in iMODFIT (48) where all the dihedral angles were fixed and only rotation and translation of internal coordinates were allowed. The iMODFIT protocol was followed as described for dealing with huge systems on http://chaconlab.org/imodfit/faq.html. After flexible fitting of the asymmetric unit, a pseudoatomic model was generated for the whole capsid in the Oligomer generator of VIPERDB (49). No clashes were observed between neighboring subunits in the generated model.
The conformational changes and domain movements occurring on release of RNA from the capsid were quantified by calculating the component placement scores (50) for corresponding pairs of capsid proteins and individual secondary-structure elements (SSEs) between empty CVA9-integrin αvβ6 and filled CVA9-integrin αvβ6 capsids (51). In addition, the Cα root mean square deviations (RMSD) between the refined models for each corresponding protein were calculated, using Chimera (47).
Difference imaging.
Difference maps were generated using the “difference map” option in Robem (41), which subtracts one map from another after bringing the two maps to similar magnifications and pixel densities. Prior to difference mapping, the X-ray structure of CVA9 (PDB 1d4m) (12) was converted to a density map using the pdb2mrc program in EMAN with 0.113 nm/pixel sampling, 400-pixel box size, and a resolution of 9.0 Å or 10.3 Å for difference imaging with filled CVA9-integrin αvβ6 or filled CVA9, respectively (40).
Surface plasmon resonance measurements of receptor binding.
Binding kinetics and affinity were studied with surface plasmon resonance (SPR) using a Biacore 2000 (GE Healthcare, Sweden) instrument. Biacore carboxymethyl-dextran CM-4 chips (GE Healthcare, Sweden) coated by amine coupling were activated with N-hydroxysuccimide and N-ethyl-N′-(dimethylaminopropyl)carbodiimide for 8 min. Then, purified CVA9 virus in 10 mM sodium acetate buffer (pH 5.0) was injected over the surface at a flow rate of 5 μl/min, resulting in 500 to 1,000 response units (RU) of virus coupled to the activated chip surface. Nonspecific sites were quenched by the addition of 1 M ethanolamine-HCl (pH 8.5) for 7 min. Purified ectodomain of integrin αVβ6 (50, 100, or 150 nM) was injected at 10 μl/min for 5 min. All measurements were done at 25°C. The buffer used for binding measurements was 10 mM HEPES (pH 7.4)–150 mM NaCl–0.005% polysorbate–1 mM MgCl2–1 mM CaCl2. BiaEvalution 4.1 (GE Healthcare, Sweden) software was used to fit the kinetic data to different kinetic models, e.g., simple 1:1 binding, a two-state model assuming integrin conformational changes, or parallel reactions of the integrin binding to the virus. Mass transfer effects were also tested. The best fit was judged by the lowest χ2 values.
The reconstructions (icosahedral, asymmetric, and tomographic) have been deposited in the Electron Microscopy Data Bank (EMDB) with accession numbers EMD-5512, -5514, -5515, -5516, -5517, and -5519. The flexible fit of the CVA9 atomic model to the empty CVA9-integrin complex has been deposited with the Protein Data Bank in Europe (PDBe) under PDB 3J2J.
RESULTS
Structure of the CVA9-integrin complex.
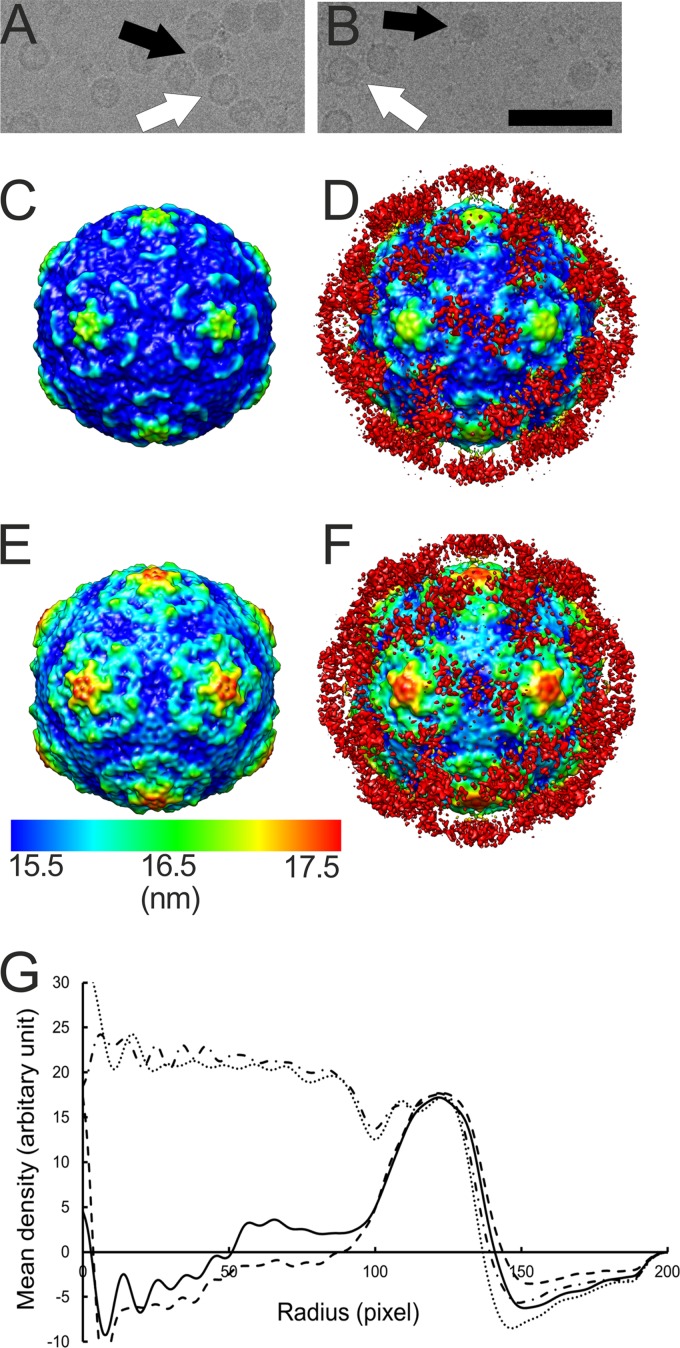
In vitro binding assays have shown integrin αvβ6 to be a high-affinity receptor for CVA9 (17, 24), which prompted us to study this interaction at the structural level. The soluble ectodomain of integrin αvβ6 was complexed with CVA9 in vitro and vitrified in order to perform cryo-EM and single-particle reconstruction, initially applying icosahedral symmetry. The micrographs indicated additional density bound to both filled and empty capsids compared to the control sample (Fig. 1A and B). The percentage of empty particles did not increase upon binding to integrin, as the control sample contained 43% empty particles (n = 1,304) compared to 37% in the integrin-bound sample (n = 835). This indicated that integrin binding to the virion did not cause RNA release under these conditions. Icosahedral reconstruction of the two capsid types (with and without integrin) showed a 2.9% increase in capsid radius of the empty capsid compared to the filled capsid whether or not integrin was bound (Fig. 1C to G). The differences in the radial profiles inside the empty capsid (below a radius of 100 nm) are due to noise (Fig. 1G). Apart from the integrin density, the capsids were directly comparable when filtered to the same resolution. This implies that binding of the integrin to filled or empty capsids did not induce any obvious changes in the virus structure (Fig. 1C to F). The icosahedrally symmetric reconstructions of the CVA9-integrin αvβ6 complexes showed very weak density for the bound integrin due to low occupancy or flexibility or both (Fig. 1C to F; red density in panels D and F). Comparison of the filled capsid-integrin reconstruction to the CVA9 atomic model indicated that the integrin binds to the virus capsid in the proximity of valine 284. This is the last residue in the atomic model of VP1. The distance between valine 284 and the integrin density is short enough for the RGD motif starting five residues later to coincide with the integrin density, close to the C terminus of VP1, between the 5-fold and 2-fold axes (Fig. 2).
Fig 1.
Electron micrographs and icosahedral reconstructions of CVA9 in complex with integrin. (A and B) Micrographs of CVA9 capsid alone (1.6-μm underfocus) and in complex with integrin αvβ6 (3.0-μm underfocus), respectively. In panels A and B, a white arrow indicates an empty particle and a black arrow indicates a filled particle. Bar, 100 nm. (C and D) Radially depth-cued isosurface representations of the filled CVA9 capsid (C) and filled CVA9 capsid-integrin αvβ6 complex (D) filtered to 10.3-Å resolution. (E and F) Radially depth-cued isosurface representations of the empty CVA9 capsid and empty CVA9 integrin αvβ6 capsid filtered to 9.9-Å resolution. In panels C to F, the view is down a 2-fold axis of symmetry. The scale bar is for the radial depth cueing. (G) Radial profiles for the empty CVA9-integrin αvβ6 complex (line with black dashes), filled CVA9-integrin αvβ6 complex (line with black dashes and dots), empty CVA9 capsid (solid line), and filled CVA9 (dotted line) (one pixel is 1.13 Å).
Fig 2.
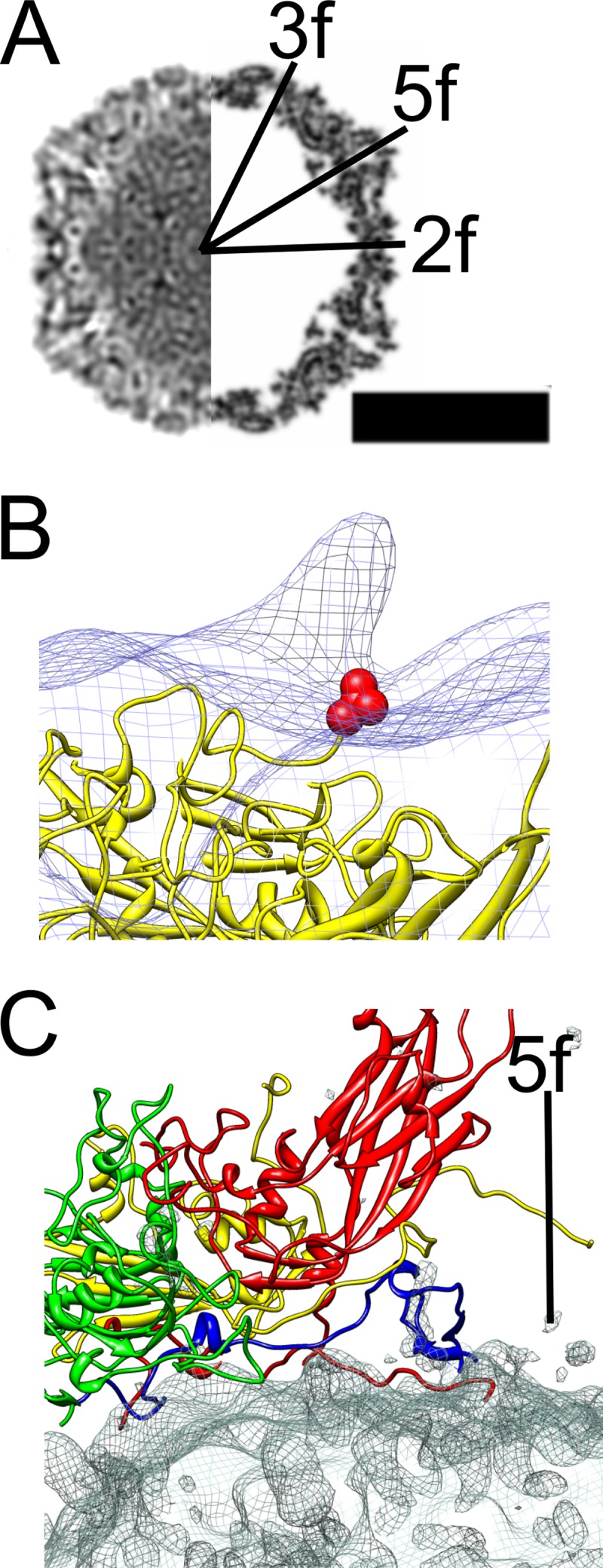
Comparison of the atomic model of CVA9 with the CVA9-integrin complex model. (A) Central cross-section view of icosahedrally symmetric filled CVA9 capsid-integrin αvβ6 density (left side) and central cross-section of simulated density for the CVA9 atomic model (PDB 1d4m; right side) (12, 44), with the symmetry axes marked 3f, 2f, and 5f. Protein and RNA are shown in black. Bar, 15 nm. (B) Comparison of asymmetric reconstruction of the filled CVA9 capsid-integrin αvβ6 complex (wire net) with the CVA9 atomic model (yellow ribbon model). The integrin density seen as a protrusion in the wire net is from a position that is symmetry related with respect to the one enforced during alignment. The integrin is on top of the VP1 C terminus shown as a red ball model of valine 284. (C) The interaction of a single protomer of CVA9 (VP1, red; VP2, green; VP3, yellow; VP4, blue) with the difference map (wire net) created by subtracting the CVA9 atomic model from the icosahedrally symmetric filled CVA9 capsid-integrin αvβ6 density. The density mainly represents RNA.
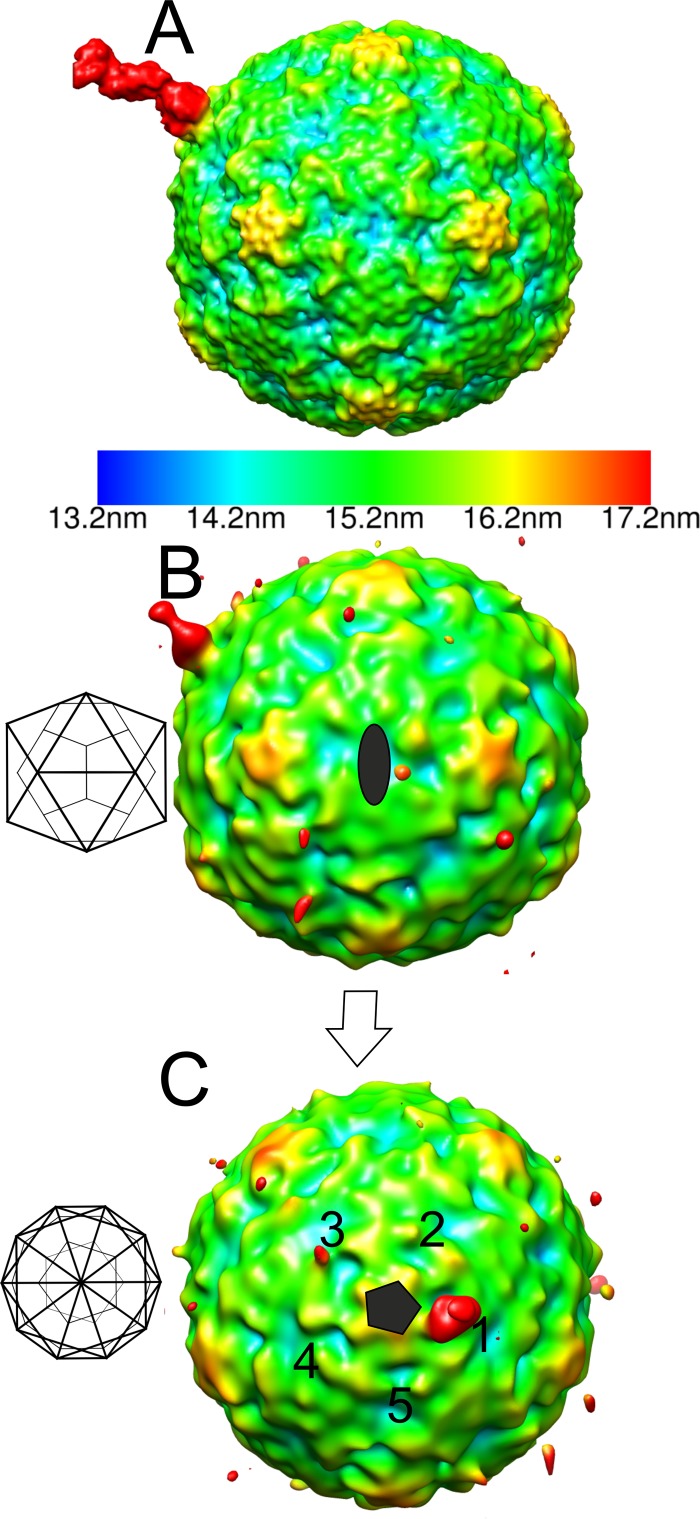
As the signal for the integrin in the icosahedral reconstructions was very weak (Table 1 and Fig. 1C to F), we also examined the number of integrins bound and the variation in conformation using cryo-ET. The results supported the hypotheses that a variable number of integrins bind the virus capsids, that they are flexible, and that there is apparently more than one integrin conformation (Fig. 3). Thus, we calculated an asymmetric reconstruction for the filled capsid-integrin complex where we utilized the previously determined icosahedral origins and orientations of the capsid to perform a limited search over 60 positions to align one integrin per capsid to the same orientation (30). We used a reference model of one β-chain of integrin αIIbβ3, bound to a filled capsid at the position adjacent to the C terminus of VP1 identified from the icosahedrally symmetric filled capsid-integrin complex reconstruction. As a result of this process, one of the integrins bound to each capsid aligned to the reference model (Fig. 4A). The remaining integrins on the capsid could be randomly distributed over the other 59 positions with an approximately equivalent signal in those 59 positions, or else the signal could be stronger than average in some positions if there is cooperative binding or steric hindrance giving rise to a preferential distribution. We found that the capsid in the asymmetric reconstruction still appeared to be approximately icosahedrally symmetric, as expected if the orientation search was successful, and there was then additional strong density on the one position biased by the reference model (Fig. 4B and C). In addition, the signal on the four equivalent positions around the same 5-fold vertex was weakest in the positions closest to that of the model integrin binding site (positions 2 and 5 compared to positions 3 and 4; Fig. 4C). This indicated that integrin binding to the capsid sterically hindered the binding of other integrins on either side (positions 2 and 5; Fig. 4C). Using both chains for the integrin changed the appearance of the integrin on position 1 (indicating model bias), but did not significantly increase the signal on the other vertices. However, when a model was used with the integrin incorrectly placed on a 5-fold vertex (“incorrect model”), the integrin density was not as strong as that seen with the correctly placed model. In addition, we calculated the pixel intensity within 59 probable receptor binding sites, excluding the biased site where we had put the integrin on the capsid surface in asymmetric reconstructions from both correct and incorrect models. After scaling of the capsid density, the pixel intensity was found to be higher in the reconstruction from the correct model than in the incorrect one (3,216 versus 2,847), implying that alignment of the remainder of the receptors is more efficient when a correct model is used. Hence, we can show probable steric hindrance in the attachment of integrin; however, we could not model the conformation of the integrin bound using this method.
Fig 3.
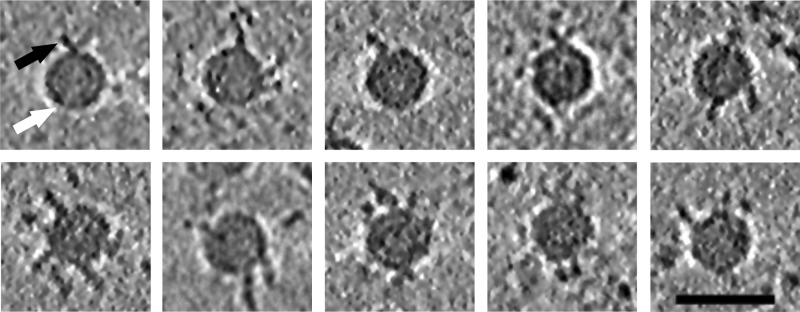
Tomographic reconstructions of the filled CVA9 capsid-integrin αvβ6 complex. Clear integrin density bound to virus particles can be seen in 0.76-nm-thick slices through the tomographic reconstructions. The number of integrins bound per capsid appears variable. Different conformations of integrin are seen attached to the capsids. White arrow, capsid; black arrow, integrin. Bar, 50 nm.
Fig 4.
Radially depth-cued isosurface representations of the asymmetric reconstruction of the filled CVA9-integrin αvβ6 capsid. (A) The reference model used in an asymmetric reconstruction run in AUTO3DEM (41) viewed down a 2-fold axis of symmetry. The reference model has an integrin β-chain in the active state bound to one position adjacent to the C terminus of VP1 on the filled CVA9 capsid reconstruction. (B) The asymmetric reconstruction generated by AUTO3DEM at a 0.5 standard deviation (SD) above the mean, showing the low occupancy of the integrin in the symmetry-related positions on the capsid, where the highest signal is for the position aligned to the modeled integrin shown in panel A. The 2-fold orientation of the capsid is shown as a schematic diagram, which is then rotated to a 5-fold orientation. (C) Five-fold view of the model described for panel B with 5 equivalent positions around one 5-fold vertex marked with numbers 1 to 5. Position 1 is that enforced by the model. Position 3 has a weak signal for the integrin. (A to C) The scale bar for the radial depth cueing is shown in panel A, chosen so that the density at the radii of the integrin is shown in red.
Measurements of receptor binding to CVA9.
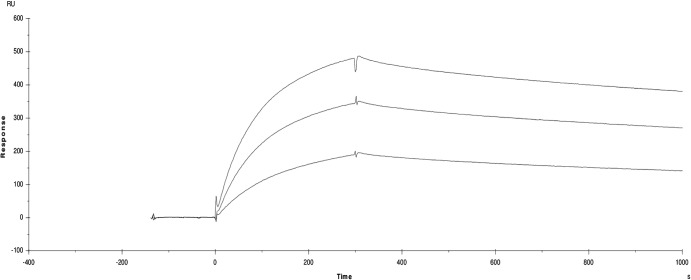
SPR measurements showed that the surface-coupled CVA9 (ligand) binds to the αvβ6 ectodomain (analyte) with nanomolar affinity (Fig. 5 and Table 2). The data could be modeled either according to a two-state model or as a parallel reaction model (Table 2), as they both appropriately fit the data at the different concentrations used (χ2 = 1 to 5; Table 2). This is in contrast to a model of 1:1 binding, which gives χ2 = 100 to 500. The two-state binding assumes a conformational change of the analyte on binding, and the parallel reaction model assumes that there are two populations of the analyte which can bind the virus with different affinities or two different binding sites on the virus. The reconstructions indicate that the binding sites on the virus are similar for the filled and empty capsids, and that this is really restricted to the flexible, exposed C terminus of VP1. Hence, it is the conformational variation in the analyte that is most likely the explanation for the fact that the simple 1:1 binding model is inappropriate for analysis of this interaction.
Fig 5.
Surface plasmon resonance sensograms for αvβ6 binding to CVA9. Data are shown for 50, 100, and 150 nM integrin injected over the CVA9 coated chip surface. The x axis is time in seconds, the y axis is the surface plasmon resonance signal in response units (RU).
Table 2.
Kinetic constants and fit for a parallel reaction model for integrin αvβ6 binding to CVA9 and for a two-state reaction model
| ka1 (1/ms) | kd1 (1/s) | ka2 (1/ms) | kd2 (1/s) | Kd1 (nM) | Kd2 (nM) | χ2 |
|---|---|---|---|---|---|---|
| Value(s) for parallel reaction model for integrin αvβ6 binding to CVA9 | ||||||
| 7.52 × 104 ± 4.8 × 104 | 3.31 × 10−5 ± 0.35 × 10−5 | 8.67 × 104 ± 3.3 × 104 | 6.55 × 10−4 ± 0.1 × 10−4 | 0.38 ± 0.24 | 7.5 ± 3.8 | 1.24–5.46 |
| ka1 (1/ms) | kd1 (1/s) | ka2 (1/s) | kd2 (1/s) | Kd (nM) | χ2 |
|---|---|---|---|---|---|
| Value(s) for two-state reaction model for integrin αvβ6 binding to CVA9 | |||||
| 8.09 × 104 ± 0.9 × 104 | 2.96 × 10−4 ± 0.3 ×10−4 | 2.46 × 10−4 ± 0.4 ×10−4 | 5.56 × 10−5 ± 0.4 ×10−5 | 0.85 ± 0.26 | 1.12–5.95 |
It is well known that integrins undergo a conformational change upon activation and ligand binding (52). Hence, the two-state model predicts a low rate of binding of the first-stage complex (on rate, ka1) which converts into a nondissociable complex slowly but which then converts back to an actively dissociating complex (Table 2). Overall, this gives a high affinity of ca. Kd = 1 nM (dissociation constant) for the receptor-virus interaction, with the rate of formation of the nondissociable complex being low (on rate, ka2) and with an even lower rate of dissociation (kd2 = 5.5 × 10−5 s−1), resulting in a very low overall off rate (Fig. 5). In contrast, the parallel-reaction model simply predicts two different binding sites or two binding modes with differing affinities (Table 2) with similar on rates but with one with a lower off rate (3.31 × 10−5 s−1 versus 6.55 × 10−4 s−1). When the ligand and the analyte were reversed, the virus bound irreversibly to the integrin coated surface and it was not possible to regenerate the surface (data not shown). Hence, we could not directly measure avidity.
Conformational changes in capsid on RNA release.
The RNA genome appears ordered in CVA9, closely contacting the inner surface of the capsid. Difference imaging of the atomic model of filled CVA9 capsids (PDB 1d4m) (12) from the icosahedrally symmetric filled CVA9 capsid-integrin αvβ6 reconstruction (Fig. 2A and C) allowed us to identify points of contact of the capsid proteins with the genome. The most obvious contacts are the N and C termini of VP4 and the N terminus of VP1, which are present below the 5-fold vertex (Fig. 2C). In addition, we confirmed that VP2 Trp38 also contacts the RNA (Fig. 2C) (12).
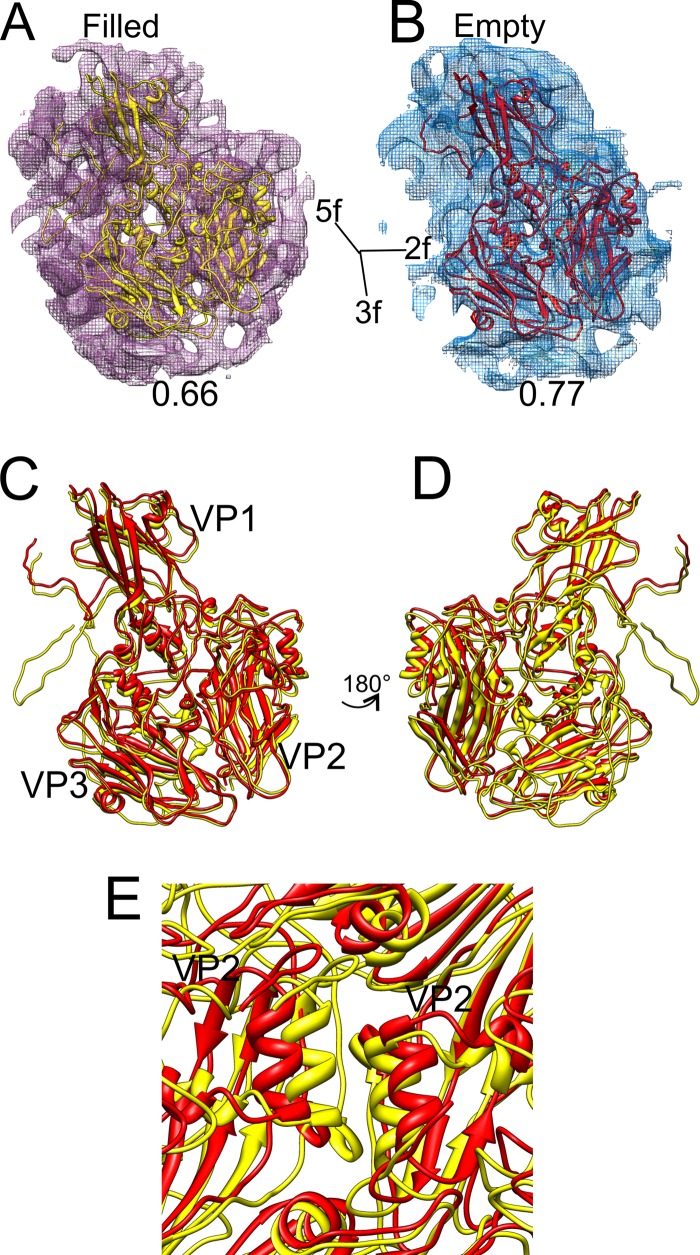
By fitting the X-ray structure of CVA9 to the integrin-bound filled capsid and by using flexible fitting to the empty capsid icosahedrally symmetric reconstructions with and without integrin (Fig. 6A and B), we were able to compare two different states of the capsid, a genome-filled compact form and the expanded empty capsid state. In contrast to the filled capsid, no density for either VP4 or the N terminus of VP1 was observed in the empty CVA9-integrin reconstruction. The protomer undergoes a translation of 3.8 Å and counterclockwise rotation of 2.8° from the RNA-filled state to the empty state (Table 3). This rotation is similar to that seen with enterovirus 71 (EV71), where it was reported to be 5.4° in a counterclockwise direction upon RNA release (53). Within the protomer, all of the three major capsid proteins, VP1, VP2, and VP3, underwent changes in orientation and translation, although in VP1 the movements were primarily restricted to translations (Table 3 and Fig. 6C and D). On inspecting the conformational changes of individual secondary-structure elements, the most evident changes observed are the translation of the VP1 β-barrel and rotation of the β-barrels of VP2 and VP3 (Table 4 and Fig. 6C and D). Such changes within the protomer tear apart neighboring G81-Y89 α-helices of adjacent VP2s at the 2-fold axis (Fig. 6E). This separation of adjacent protomers has been previously observed for VP2 helices 91 to 98 of coxsackievirus A7 (51) and VP2 αA helices of enterovirus 71 (53) and human rhinovirus 2 (54). The conformational changes occurring in the capsid after RNA release are similar in the capsids with and without integrins, so those changes are not induced by integrin binding.
Fig 6.
Flexible fitting of the atomic model of CVA9 into the density map representing both the filled and empty integrin-bound capsids. (A and B) Atomic structure of CVA9 protomer (VP1, VP2, and VP3) shown fitted in a single asymmetric unit of a filled CVA9 integrin αvβ6 capsid density map and empty CVA9 integrin αvβ6 capsid density map (wire net), respectively, with the correlation value given below each map. (A and B) The density maps are shown at 2.5 standard deviations above the mean. The direction of the symmetry axes is shown. (C) Superimposed models of filled (yellow) and empty (red) fitted structures. Panel D presents a view rotated 180° with respect to panel C. Symmetry axes are marked in panels C and D. Panel E shows the G81-Y89 α-helices of neighboring VP2 moving apart in the empty fitted structure compared to the filled one, around a 2-fold axis. In panels A to E, all the atomic models are shown in ribbon form.
Table 3.
Component placement score and Cα RMSD for the individual proteins between empty and filled capsids (50)
| Protein name | Translation (Å) | Rotation (°) | Cα RMSD, empty to full (Å) |
|---|---|---|---|
| Protomer (VP1, 63–284; VP2, 1–238; VP3, 1–252) | 3.8 | 2.8 | 4.04 |
| VP1 (63–284) | 4.6 | 1.0 | 4.67 |
| VP2 (1–238) | 3.4 | 4.4 | 3.58 |
| VP3 (1–252) | 3.6 | 4.2 | 3.89 |
Table 4.
Component placement scores for individual secondary-structure elements between filled and empty capsids (50)
| Protein name | Residue range(s) | Secondary-structure-element type | Translation (Å) | Rotation (°) |
|---|---|---|---|---|
| VP1 | 64–68 | α-Helix | 4.7 | 1.0 |
| 101–109 | α-Helix | 4.6 | 1.0 | |
| 160–163 | α-Helix | 4.6 | 1.0 | |
| 210–213 | α-Helix | 4.4 | 1.0 | |
| 72–80, 116–127, 179–183, 232–243 | β-Sheet | 4.8 | 1.0 | |
| 90–94, 141–147, 169–173, 218–223 | β-Sheet | 4.7 | 1.0 | |
| 112–114, 246–248 | β-Sheet | 4.5 | 1.0 | |
| VP2 | 81–89 | α-Helix | 4.1 | 4.4 |
| 170–175 | α-Helix | 3.2 | 4.4 | |
| 60–62, 96–102, 188–193, 230–239 | β-Sheet | 3.0 | 4.4 | |
| 69–73, 112–119, 178–182, 210–218 | β-Sheet | 3.1 | 4.4 | |
| 5–9, 12–16 | β-Sheet | 2.7 | 4.4 | |
| VP3 | 43–47 | α-Helix | 4.3 | 4.2 |
| 99–104 | α-Helix | 4.0 | 4.2 | |
| 145–148 | α-Helix | 1.9 | 4.2 | |
| 81–86, 129–135, 152–157, 189–194 | β-Sheet | 2.5 | 4.2 | |
| 70–73, 114–120, 163–168, 207–216 | β-Sheet | 2.9 | 4.2 | |
| 109–111, 221–223 | β-Sheet | 4.0 | 4.2 |
DISCUSSION
A prerequisite for virus entry into a cell is that it must bind to the cell surface and subsequently trigger uptake by the cell. Increasingly, it has been observed that this is a multistep process, involving many factors (55, 56). In CVA9, one of the first steps of cell recognition is the binding to an integrin receptor (24). We showed that the Kd of this reaction with the integrin αvβ6 is in the low nanomolar range (Table 2) and that dissociation occurs through interaction with a site on the viral capsid close to the predicted RGD loop of VP1 (Fig. 1, 2, and 4) based on the atomic model of the capsid (12). As there are 60 such equivalent sites on the capsid, we could show that there is apparent steric hindrance preventing occupancy of all sites under the conditions used with an ∼200-fold molar excess of integrin over the capsid (Fig. 4). As the percentages of empty particles with or without integrin were found to be similar, integrin binding did not induce uncoating of RNA, which is similar to observations made for human parechovirus 1 binding to both αvβ6 and αvβ3 (28).
The reconstructions of the CVA9 empty particle most likely represent the last stage of the uncoating process. We see the structural rearrangements produced in the viral capsid by the loss of VP4 and the delivery of the genomic RNA, independently of integrin binding (Fig. 1 and 6 and Tables 3 and 4). Further, the rotations and translations of the capsid proteins result in a more porous structure (Fig. 2) defined in more detail by flexible fitting from a known atomic model of the filled capsid into the empty capsid. We propose that RNA exits through pores that are produced between adjacent copies of VP2 at or close to the 2-fold axes as has recently been suggested for four other picornaviruses: coxsackievirus A7 (51), enterovirus 71 (53), poliovirus (57), and human rhinovirus 2 (54). We have analyzed two end states; results from analysis of poliovirus in the act of releasing its RNA are also in line with these observations (57).
Here we used a number of different approaches to understand the integrin-capsid interactions. The icosahedrally symmetric reconstruction allowed us to pinpoint the position of the integrins on the capsid surface, but not to address their conformation, due to the incoherent averaging of the bound integrin attached to flexible RGD loops (23, 28). The complementary approaches of electron cryo-tomography and asymmetric reconstruction underlined the heterogeneity of the integrins seen bound to the capsid. In the cell membrane, the integrin conformation may be more constrained than in our experiments with soluble integrin. The integrin heterogeneity was reflected in the clearly bipartite kinetics of integrin attachment to CVA9 seen in SPR measurements, which showed a faster and a slower phase on binding or a higher and a lower off rate, depending on the model (Fig. 5 and Table 2). This is because we could model the SPR binding kinetics as either a two-state or a parallel-state reaction. As the integrin binding sites on the empty and full capsids are similar, and the RGD loop is surface exposed and highly mobile, we suggest that the kinetics are mainly influenced by the existence of different conformations of the integrin (52). As the two kinetic models are equally plausible, we remain with two hypotheses, one that suggests that virus binding induces a conformational change in the integrin, and the alternative, that there are two integrin populations that are released from the capsid with different off rates. In terms of the infection process in the cell, there are advantages for both the models. If the virus can bind any of the three conformations of the integrin, then the probability of binding a receptive host cell is increased, thus increasing the probability of infection. This would be the case if the model of the parallel-state reaction is correct. This is the model we favor. If only one integrin conformation can be bound, then there are fewer potential binding partners per cell, but if virus binding then immediately triggered a conformational change (two-state model) leading to integrin signaling and uptake, the probability of entry at this second step would be improved. Echovirus 1, for instance, which binds to the bent α2β1 integrin conformation, causes integrin clustering, probably mediating signaling for endocytosis, and at later time points colocalizes with internalized integrin (15, 25, 58). It is likely that avidity plays a role in the in vivo situation, resulting in an even higher overall affinity of the virus for the cell surface. The slow dissociation of the 1:1 complex in SPR indicates that once the virus is bound, it is anchored on the cell surface very tightly, in a waiting mode for the triggering of the internalization step. Independent support for the model of the parallel-state reaction comes from published work showing that in infected A549 cells, CVA9 does not colocalize with internalized integrin, suggesting that integrin signaling does not promote CVA9 internalization. Instead, other molecules such as β2-microglobulin and GRP78 are probably required for internalization (15, 26).
In conclusion, our results shed light on the structural basis of integrin binding to CVA9, and on the molecular events leading to CVA9 cell entry and uncoating.
ACKNOWLEDGMENTS
We thank Ritva Kajander, Turku University, Hilkka Lankinen and Carl-Mikael Suomivuori, Helsinki University, for excellent technical assistance, Giovanni Cardone, UCSD, for active discussion and support with programming, and J. R. Lopez Blanco, CSIC, for technical support on the usage of the iMODFIT software. The Biocenter Finland National Cryo Electron Microscopy Unit, Institute of Biotechnology, Helsinki University, and the CSC-IT Center for Science Ltd. kindly provided facilities.
This work was supported by the Academy of Finland Centre of Excellence Programme in Virus Research (2006 to 2011; 1129684 to S.J.B.), Academy of Finland (1139178 to S.J.B.; 128539 to P.S.; and 122215 to T.H.), Sigrid Juselius Foundation (S.J.B. and T.H.), the National Graduate School in Informational and Structural Biology (J.J.T.S.), and the Helsinki Graduate Program in Biotechnology and Molecular Biology (S.S.).
Footnotes
Published ahead of print 30 January 2013
REFERENCES
- 1. Grist NR, Bell EJ, Assaad F. 1978. Enteroviruses in human disease. Prog. Med. Virol. 24:114–157 [PubMed] [Google Scholar]
- 2. Michos AG, Syriopoulou VP, Hadjichristodoulou C, Daikos GL, Lagona E, Douridas P, Mostrou G, Theodoridou M. 2007. Aseptic meningitis in children: analysis of 506 cases. PLoS One 2:e674 doi:10.1371/journal.pone.0000674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Whitton JL, Cornell CT, Feuer R. 2005. Host and virus determinants of picornavirus pathogenesis and tropism. Nat. Rev. Microbiol. 3:765–776 [DOI] [PubMed] [Google Scholar]
- 4. Cooper LT., Jr 2009. Myocarditis. N. Engl. J. Med. 360:1526–1538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Eisenhut M, Algawi B, Wreghitt T, Foweraker J, McKee T, Miles R, Challener J. 2000. Fatal coxsackie A9 virus infection during an outbreak in a neonatal unit. J. Infect. 40:297–298 [DOI] [PubMed] [Google Scholar]
- 6. Blomqvist S, Paananen A, Savolainen-Kopra C, Hovi T, Roivainen M. 2008. Eight years of experience with molecular identification of human enteroviruses. J. Clin. Microbiol. 46:2410–2413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cui A, Yu D, Zhu Z, Meng L, Li H, Liu J, Liu G, Mao N, Xu W. 2010. An outbreak of aseptic meningitis caused by coxsackievirus A9 in Gansu, the People's Republic of China. Virol. J. 7:72 doi:10.1186/1743-422X-7-72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Clements GB, Galbraith DN, Taylor KW. 1995. Coxsackie B virus infection and onset of childhood diabetes. Lancet 346:221–223 [DOI] [PubMed] [Google Scholar]
- 9. Roivainen M, Knip M, Hyöty H, Kulmala P, Hiltunen M, Vahasalo P, Hovi T, Åkerblom HK. 1998. Several different enterovirus serotypes can be associated with prediabetic autoimmune episodes and onset of overt IDDM. J. Med. Virol. 56:74–78 [DOI] [PubMed] [Google Scholar]
- 10. Hiltunen M, Hyöty H, Knip M, Ilonen J, Reijonen H, Vahasalo P, Roivainen M, Lönnrot M, Leinikki P, Hovi T, Åkerblöm HK, and the Childhood Diabetes in Finland (DiMe) Study Group 1997. Islet cell antibody seroconversion in children is temporally associated with enterovirus infections. J. Infect. Dis. 175:554–560 [DOI] [PubMed] [Google Scholar]
- 11. Hyöty H, Hiltunen M, Knip M, Laakkonen M, Vahasalo P, Karjalainen J, Koskela P, Roivainen M, Leinikki P, Hovi T, Åkerblom HK. 1995. A prospective study of the role of coxsackie B and other enterovirus infections in the pathogenesis of IDDM. Diabetes 44:652–657 [DOI] [PubMed] [Google Scholar]
- 12. Hendry E, Hatanaka H, Fry E, Smyth M, Tate J, Stanway G, Santti J, Maaronen M, Hyypiä T, Stuart D. 1999. The crystal structure of coxsackievirus A9: new insights into the uncoating mechanisms of enteroviruses. Structure 7:1527–1538 [DOI] [PubMed] [Google Scholar]
- 13. Chang KH, Auvinen P, Hyypiä T, Stanway G. 1989. The nucleotide sequence of coxsackievirus A9; implications for receptor binding and enterovirus classification. J. Gen. Virol. 70:3269–3280 [DOI] [PubMed] [Google Scholar]
- 14. Muckelbauer JK, Kremer M, Minor I, Tong L, Zlotnick A, Johnson JE, Rossmann MG. 1995. Structure determination of coxsackievirus B3 to 3.5 Å resolution. Acta Crystallogr. D Biol. Crystallogr. 51:871–887 [DOI] [PubMed] [Google Scholar]
- 15. Heikkilä O, Susi P, Tevaluoto T, Harma H, Marjomäki V, Hyypiä T, Kiljunen S. 2010. Internalization of coxsackievirus A9 is mediated by {beta}2-microglobulin, dynamin, and Arf6 but not by caveolin-1 or clathrin. J. Virol. 84:3666–3681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Roivainen M, Piirainen L, Hovi T, Virtanen I, Riikonen T, Heino J, Hyypiä T. 1994. Entry of coxsackievirus A9 into host cells: specific interactions with alpha v beta 3 integrin, the vitronectin receptor. Virology 203:357–365 [DOI] [PubMed] [Google Scholar]
- 17. Williams CH, Kajander T, Hyypiä T, Jackson T, Sheppard D, Stanway G. 2004. Integrin alpha v beta 6 is an RGD-dependent receptor for coxsackievirus A9. J. Virol. 78:6967–6973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Santti J, Harvala H, Kinnunen L, Hyypiä T. 2000. Molecular epidemiology and evolution of coxsackievirus A9. J. Gen. Virol. 81:1361–1372 [DOI] [PubMed] [Google Scholar]
- 19. Hughes PJ, Horsnell C, Hyypiä T, Stanway G. 1995. The coxsackievirus A9 RGD motif is not essential for virus viability. J. Virol. 69:8035–8040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Takagi J, Petre BM, Walz T, Springer TA. 2002. Global conformational rearrangements in integrin extracellular domains in outside-in and inside-out signaling. Cell 110:599–611 [DOI] [PubMed] [Google Scholar]
- 21. Nishida N, Xie C, Shimaoka M, Cheng Y, Walz T, Springer TA. 2006. Activation of leukocyte beta2 integrins by conversion from bent to extended conformations. Immunity 25:583–594 [DOI] [PubMed] [Google Scholar]
- 22. Xiong JP, Stehle T, Zhang R, Joachimiak A, Frech M, Goodman SL, Arnaout MA. 2002. Crystal structure of the extracellular segment of integrin alpha Vbeta3 in complex with an Arg-Gly-Asp ligand. Science 296:151–155 [DOI] [PubMed] [Google Scholar]
- 23. Lindert S, Silvestry M, Mullen TM, Nemerow GR, Stewart PL. 2009. Cryo-electron microscopy structure of an adenovirus-integrin complex indicates conformational changes in both penton base and integrin. J. Virol. 83:11491–11501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Heikkilä O, Susi P, Stanway G, Hyypiä T. 2009. Integrin alphaVbeta6 is a high-affinity receptor for coxsackievirus A9. J. Gen. Virol. 90:197–204 [DOI] [PubMed] [Google Scholar]
- 25. Xing L, Huhtala M, Pietiäinen V, Käpylä J, Vuorinen K, Marjomäki V, Heino J, Johnson MS, Hyypiä T, Cheng RH. 2004. Structural and functional analysis of integrin alpha2I domain interaction with echovirus 1. J. Biol. Chem. 279:11632–11638 [DOI] [PubMed] [Google Scholar]
- 26. Triantafilou M, Triantafilou K, Wilson KM, Takada Y, Fernandez N, Stanway G. 1999. Involvement of beta2-microglobulin and integrin alphavbeta3 molecules in the coxsackievirus A9 infectious cycle. J. Gen. Virol. 80:2591–2600 [DOI] [PubMed] [Google Scholar]
- 27. Chiu CY, Mathias P, Nemerow GR, Stewart PL. 1999. Structure of adenovirus complexed with its internalization receptor, alphavbeta5 integrin. J. Virol. 73:6759–6768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Seitsonen J, Susi P, Heikkilä O, Sinkovits RS, Laurinmäki P, Hyypiä T, Butcher SJ. 2010. Interaction of alphaVbeta3 and alphaVbeta6 integrins with human parechovirus 1. J. Virol. 84:8509–8519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hafenstein S, Palermo LM, Kostyuchenko VA, Xiao C, Morais MC, Nelson CD, Bowman VD, Battisti AJ, Chipman PR, Parrish CR, Rossmann MG. 2007. Asymmetric binding of transferrin receptor to parvovirus capsids. Proc. Natl. Acad. Sci. U. S. A. 104:6585–6589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Parent KN, Gilcrease EB, Casjens SR, Baker TS. 2012. Structural evolution of the P22-like phages: comparison of Sf6 and P22 procapsid and virion architectures. Virology 427:177–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Casasnovas JM, Springer TA. 1995. Kinetics and thermodynamics of virus binding to receptor. Studies with rhinovirus, intercellular adhesion molecule-1 (ICAM-1), and surface plasmon resonance. J. Biol. Chem. 270:13216–13224 [DOI] [PubMed] [Google Scholar]
- 32. Jackson T, Sheppard D, Denyer M, Blakemore W, King AM. 2000. The epithelial integrin alphavbeta6 is a receptor for foot-and-mouth disease virus. J. Virol. 74:4949–4956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bergelson JM, Shepley MP, Chan BM, Hemler ME, Finberg RW. 1992. Identification of the integrin VLA-2 as a receptor for echovirus 1. Science 255:1718–1720 [DOI] [PubMed] [Google Scholar]
- 34. Nelsen-Salz B, Eggers HJ, Zimmermann H. 1999. Integrin alpha(v)beta3 (vitronectin receptor) is a candidate receptor for the virulent echovirus 9 strain Barty. J. Gen. Virol. 80:2311–2313 [DOI] [PubMed] [Google Scholar]
- 35. Triantafilou K, Triantafilou M, Takada Y, Fernandez N. 2000. Human parechovirus 1 utilizes integrins alphavbeta3 and alphavbeta1 as receptors. J. Virol. 74:5856–5862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Baker TS, Olson NH, Fuller SD. 1999. Adding the third dimension to virus life cycles: three-dimensional reconstruction of icosahedral viruses from cryo-electron micrographs. Microbiol. Mol. Biol. Rev. 63:862–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mastronarde DN. 2005. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152:36–51 [DOI] [PubMed] [Google Scholar]
- 38. Mindell JA, Grigorieff N. 2003. Accurate determination of local defocus and specimen tilt in electron microscopy. J. Struct. Biol. 142:334–347 [DOI] [PubMed] [Google Scholar]
- 39. Kivioja T, Ravantti J, Verkhovsky A, Ukkonen E, Bamford D. 2000. Local average intensity-based method for identifying spherical particles in electron micrographs. J. Struct. Biol. 131:126–134 [DOI] [PubMed] [Google Scholar]
- 40. Ludtke SJ, Baldwin PR, Chiu W. 1999. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 128:82–97 [DOI] [PubMed] [Google Scholar]
- 41. Yan X, Sinkovits RS, Baker TS. 2007. AUTO3DEM—an automated and high throughput program for image reconstruction of icosahedral particles. J. Struct. Biol. 157:73–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yan X, Dryden KA, Tang J, Baker TS. 2007. Ab initio random model method facilitates 3D reconstruction of icosahedral particles. J. Struct. Biol. 157:211–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. van Heel M, Harauz G. 1986. Resolution criteria for three dimensional reconstruction. Optik 73:119–122 [Google Scholar]
- 44. Heymann JB. 2001. Bsoft: image and molecular processing in electron microscopy. J. Struct. Biol. 133:156–169 [DOI] [PubMed] [Google Scholar]
- 45. Kremer JR, Mastronarde DN, McIntosh JR. 1996. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116:71–76 [DOI] [PubMed] [Google Scholar]
- 46. Zhu J, Luo BH, Xiao T, Zhang C, Nishida N, Springer TA. 2008. Structure of a complete integrin ectodomain in a physiologic resting state and activation and deactivation by applied forces. Mol. Cell 32:849–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25:1605–1612 [DOI] [PubMed] [Google Scholar]
- 48. Lopez-Blanco JR, Garzon JI, Chacon P. 2011. iMod: multipurpose normal mode analysis in internal coordinates. Bioinformatics 27:2843–2850 [DOI] [PubMed] [Google Scholar]
- 49. Shepherd CM, Borelli IA, Lander G, Natarajan P, Siddavanahalli V, Bajaj C, Johnson JE, Brooks CL, Reddy VS. 2006. VIPERdb: a relational database for structural virology. Nucleic Acids Res. 34:D386–D389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Topf M, Lasker K, Webb B, Wolfson H, Chiu W, Sali A. 2008. Protein structure fitting and refinement guided by cryo-EM density. Structure 16:295–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Seitsonen JJ, Shakeel S, Susi P, Pandurangan AP, Sinkovits RS, Hyvonen H, Laurinmaki P, Yla-Pelto J, Topf M, Hyypia T, Butcher SJ. 2012. Structural analysis of coxsackievirus A7 reveals conformational changes associated with uncoating. J. Virol. 86:7207–7215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Arnaout MA, Goodman SL, Xiong JP. 2007. Structure and mechanics of integrin-based cell adhesion. Curr. Opin. Cell Biol. 19:495–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wang X, Peng W, Ren J, Hu Z, Xu J, Lou Z, Li X, Yin W, Shen X, Porta C, Walter TS, Evans G, Axford D, Owen R, Rowlands DJ, Wang J, Stuart DI, Fry EE, Rao Z. 2012. A sensor-adaptor mechanism for enterovirus uncoating from structures of EV71. Nat. Struct. Mol. Biol. 19:424–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Garriga D, Pickl-Herk A, Luque D, Wruss J, Caston JR, Blaas D, Verdaguer N. 2012. Insights into minor group rhinovirus uncoating: the X-ray structure of the HRV2 empty capsid. PLoS Pathog. 8:e1002473 doi:10.1371/journal.ppat.1002473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Mercer J, Helenius A. 2012. Gulping rather than sipping: macropinocytosis as a way of virus entry. Curr. Opin. Microbiol. 15:490–499 [DOI] [PubMed] [Google Scholar]
- 56. Mercer J, Schelhaas M, Helenius A. 2010. Virus entry by endocytosis. Annu. Rev. Biochem. 79:803–833 [DOI] [PubMed] [Google Scholar]
- 57. Bostina M, Levy H, Filman DJ, Hogle JM. 2011. Poliovirus RNA is released from the capsid near a twofold symmetry axis. J. Virol. 85:776–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Jokinen J, White DJ, Salmela M, Huhtala M, Käpylä J, Sipilä K, Puranen JS, Nissinen L, Kankaanpää P, Marjomäki V, Hyypiä T, Johnson MS, Heino J. 2010. Molecular mechanism of alpha2beta1 integrin interaction with human echovirus 1. EMBO J. 29:196–208 [DOI] [PMC free article] [PubMed] [Google Scholar]