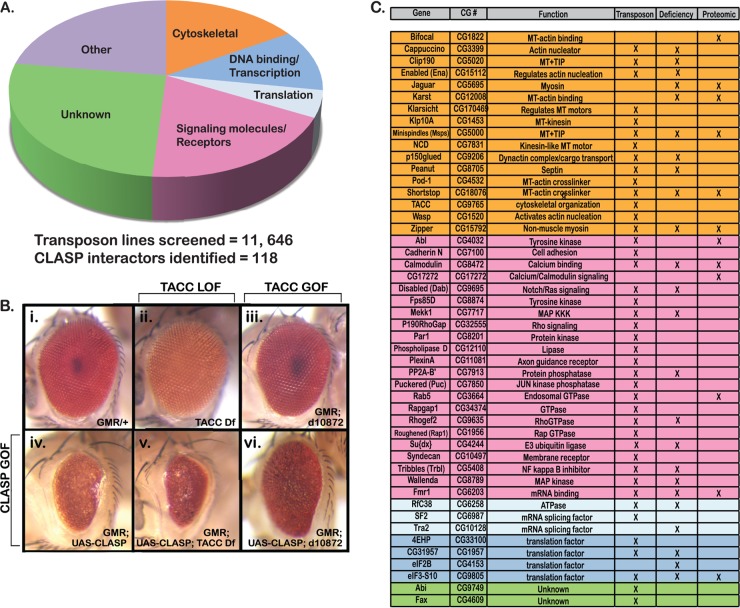
Fig 1.
Identification of CLASP interactors. The CLASP interactome was defined through in vivo genetic modifier screens using both the deletion and transposon insertion subsets of the Exelixis collection. (A) A total of 11,646 individual transposon insertions were screened to identify genes that modified the fully penetrant dominant rough eye phenotype observed upon CLASP misexpression. Overall, 118 CLASP interactors were identified. Interactors could be grouped into functional categories on the basis of gene ontology: cytoskeletal regulators, DNA binding and transcriptional regulators, translational regulators, signaling molecules and receptors, unknown function, and others. Other functions included ion transport, metabolism, and general protein binding. (B) TACC loss-of-function (LOF) (ii; TACC deficiency [Df]) and gain-of-function (GOF) (iii; d10872) mutants were used to evaluate genetic interactions with CLASP in the Drosophila eye. Loss of TACC expression enhanced the CLASP overexpression phenotype (iv and v), while overexpression of TACC suppressed the phenotype (iv and vi). (C) Forty-eight interactors were selected for further analysis in S2R+ cells on the basis of gene ontogeny and/or reported physical/functional interactions with cytoskeletal proteins (group colors correspond to the functional categories described for panel A).