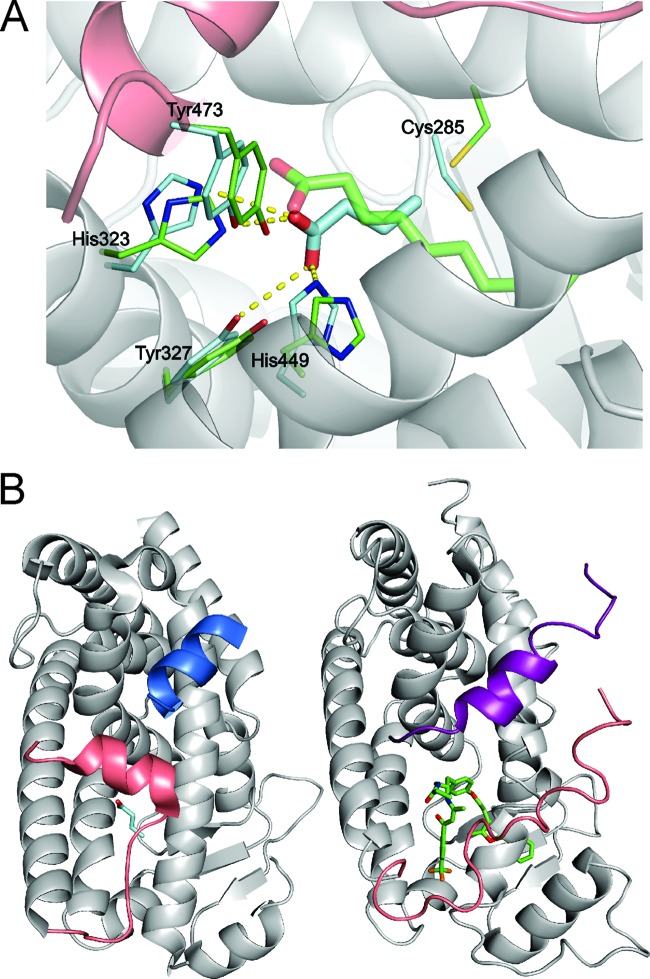
Fig 6.
Modeling of butyrate into the PPARγ binding pocket. (A) The model reveals the complex between butyrate and PPARγ with the best HADDOCK score (butyrate is shown as cyan sticks), overlaid with the crystal structure of the decanoic acid complex with PPARγ (3U9Q, a decanoic acid, is shown as green sticks) by aligning the protein backbone atoms of the two structures (ribbon displayed for the HADDOCK model). The displayed protein side chains are shown as thin cyan or green sticks and the side chains making contacts with the docked butyrate or decanoic acid, respectively. Hydrogen bond contacts between the butyrate and the protein are shown as yellow dashed lines. (B) Comparison of the binding location for coactivator peptide PGC-1α (blue ribbon) bound to PPARγ (HADDOCK model of the PPARγ-butyrate complex, left) with the binding site for the SMRT corepressor peptide (purple ribbon) binding to PPARα (1KKQ; right) (69). In both structures, the C-terminal portion of the PPAR molecule that forms helix AF-2 is colored pink for comparison. The structures were aligned using the backbone atoms of the receptors.