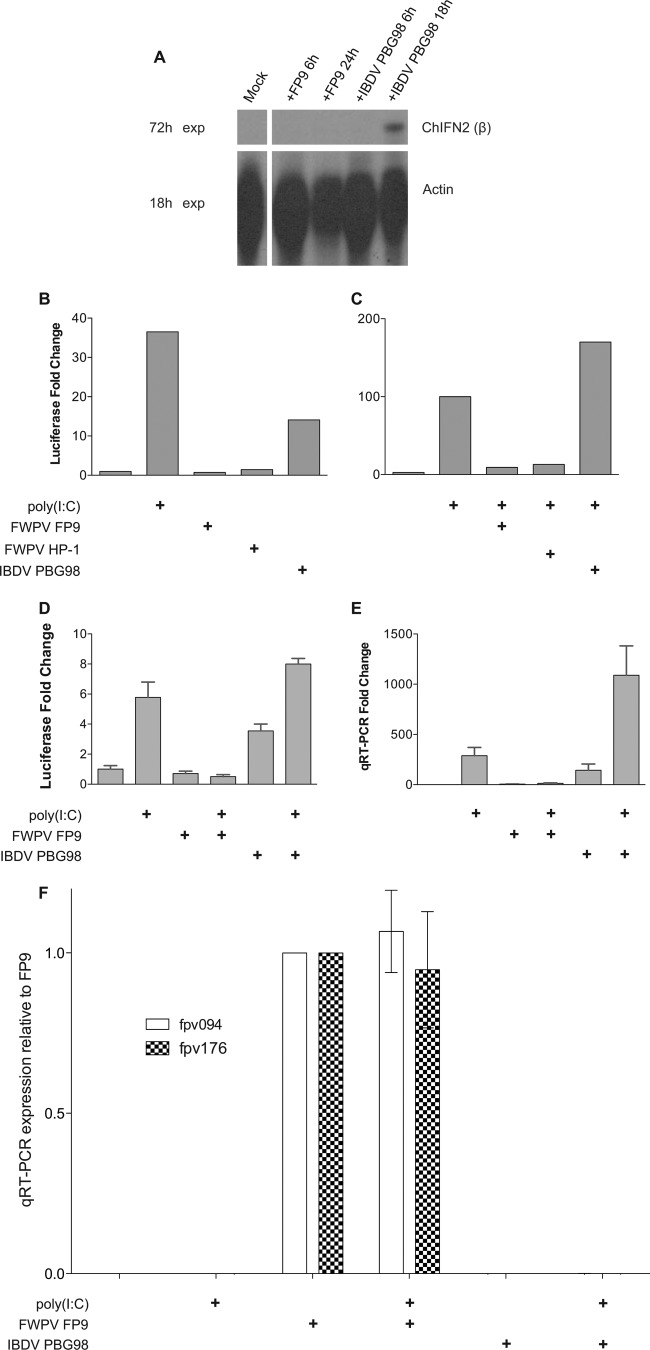
Fig 1.
Induction of the ChIFN2 promoter upon transfection with poly(I·C) and/or infection with viruses. (A) RNase protection analysis of expression of ChIFN2 in DF-1 cells upon virus infection. DF-1 cells, mock treated or infected with virus (FWPV strain FP9 or IBDV strain PBG98) for the times stated, were analyzed for expression of ChIFN2 by RNase protection using 10 μg RNA. The levels of control chicken β-actin are also shown. Gels were exposed (exp) for 18 h or 72 h. (B to D) Luciferase reporter analysis of expression of ChIFN2 in DF-1 cells. DF-1 cells were transfected with the ChIFN2 promoter luciferase reporter plasmid (pChIFN2lucter) and plasmid pJATlacZ, constitutively expressing β-galactosidase from the rat β-actin promoter (31). Following recovery for 24 h, cells were either left uninfected or infected (at an MOI of 10) with poxviruses (attenuated FWPV FP9 or its progenitor, HP1) or attenuated IBDV vaccine strain PBG98. Following infection for 4 h, cells were either left untreated or transfected with poly(I·C) (10 μg ml−1) and incubated for 16 h. Luciferase expression values were normalized to those of β-galactosidase. ChIFN2 expression levels were compared to the level of the uninduced control to calculate the fold induction. (B) Induction of the ChIFN2 promoter by transfected poly(I·C) as a positive control or by infection alone. (C) Modulation of induction of the ChIFN2 promoter mediated by transfected poly(I·C) following infection with FWPV (FP9 or HP1) or IBDV PBG98. (D and E) Modulation of induction of the ChIFN2 promoter mediated by virus infection or by transfected poly(I·C) following infection with FWPV FP9 or IBDV PBG98. Samples from the same experiment were split for luciferase assay (D) and for qRT-PCR (E). ChIFN2 expression levels were calculated relative to those of GAPDH and the untreated control. (F) The same samples used for panels D and E were tested for expression of early (fpv094) and late (fpv176) genes by qRT-PCR. Their expression was normalized against that of GAPDH and is presented relative to that in the FP9 sample.