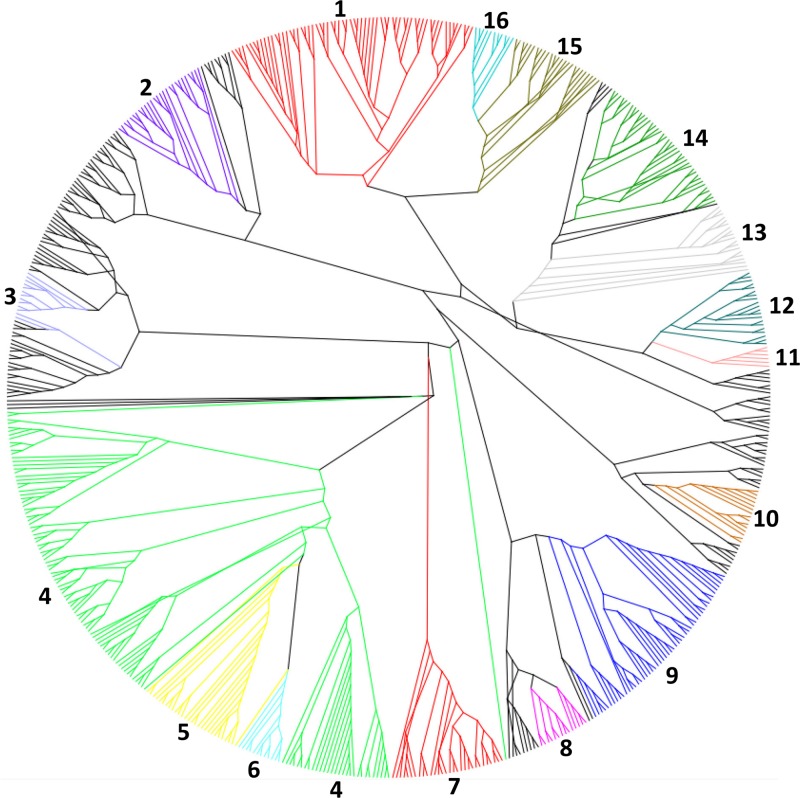
Fig 5.
Radial cladogram of bacteriophage endolysins of all characterized phages. The cladogram was constructed using PHYLIP v3.68 with a neighbor-joining algorithm. The resulting cladogram was represented and arranged, without being altered, using the FigTree software (version 1.3.1). Highlighted areas are numbered and correspond to the more-conserved clades. 1, Mycobacterium/Siphoviridae/AMI-2; 2, G+/GH25; 3, Lactococcus/Siphoviridae/AMI-2; 4, G−/LYSO; 5, G−/MURA; 6, Streptococcus/Siphoviridae/NLPC-P60; 7, G−/Podoviridae/AMI-2; 8, G−/Myoviridae/VANY; 9, Staphylococcus/Siphoviridae; 10, G−/GH19; 11, Escherichia/Tectiviridae/SLT; 12, Mycobacterium/Siphoviridae/NLPC-P60/GH25; 13, Mycobacterium/Siphoviridae/PET-M15-4/PG1; 14, Mycobacterium/Siphoviridae/PETM-15-4/GH19; 15, Mycobacterium/Siphoviridae/PET-M23/AMI-2; 16, Mycobacterium/Siphoviridae/PET-C39-2/GH19.