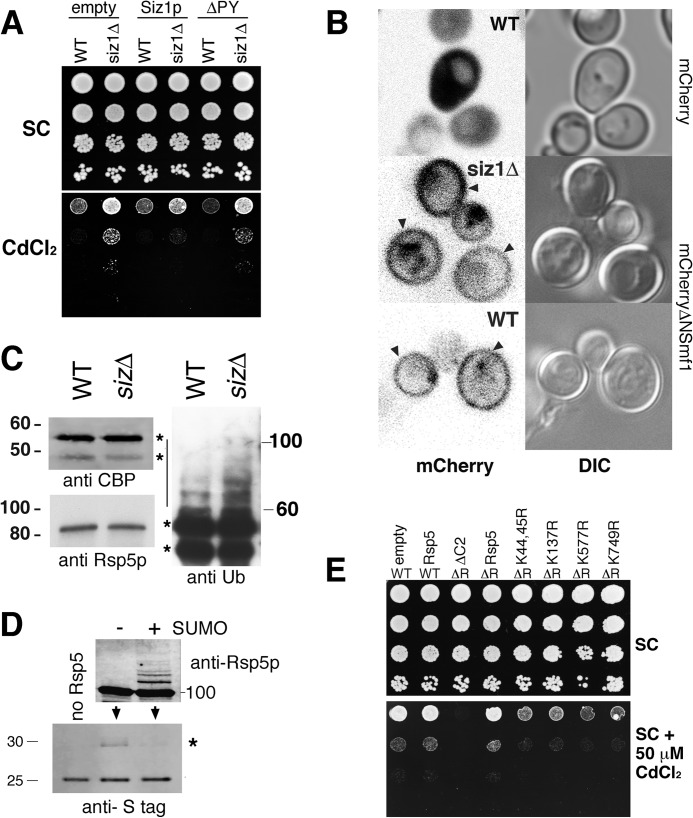
FIGURE 3.
A, wild-type yeast (WT) and a siz1Δ strain were transformed with empty FLAG (empty), FLAG-Siz1p (Siz1p), or FLAG-Siz1pΔPY (ΔPY) as indicated. Serial 10-fold dilutions of stationary phase cultures were replica-plated onto synthetic complete media without or with 50 μm CdCl2 (upper panel and lower panel, respectively) and incubated for 3 days at 30 °C. B, confocal imaging of mCherry and mCherry-tagged ΔNSmf1p and differential interference contrast (DIC) images of WT and siz1Δ yeast strains grown to log phase in metal-depleted media are shown. Arrowheads indicate mCherryΔNSmf1p signal at cell periphery. C, TAP-Bsd2p was expressed in WT yeast or siz1Δsiz2Δ strain (sizΔ) and TAP-purified. Purified proteins were subjected to SDS-PAGE followed by immunoblotting with anti-calmodulin-binding protein (CBP), anti-Rsp5, and anti-ubiquitin antibodies. Asterisks indicate cleaved (lower band) and uncleaved (upper band) forms of TAP-Bsd2p; bar indicates co-purifying HMW ubiquitin conjugates. D, upper panel shows recombinant Rsp5p (as visualized using anti-Rsp5 antibodies) subjected to in vitro SUMOylation reaction with Siz1p(1–465) (+) with a negative control that contained all the SUMOylation components except SUMO (−). Modified and unmodified Rsp5p were then used in an in vitro ubiquitylation reaction (lower panel) using the cytoplasmic portion of Bsd2p visualized using anti-Bsd2 antibodies. Asterisk indicates position of monoubiquitylated Bsd2p. E, cadmium sensitivity assays show lysine-to-arginine point mutations in SUMOylation sites identified in MS/MS analysis. Serial 10-fold dilutions were grown as in A of WT or an rsp5Δ strain (ΔR) expressing an empty plasmid, WT Rsp5 (Rsp5), an N-terminal deletion of Rsp5 that lacks the C2 domain (ΔC2) or full-length Rsp5 with lysine-to-arginine point mutations at the amino acid positions specified.