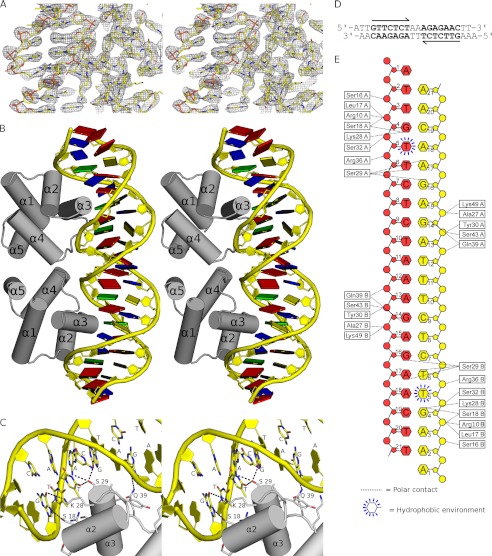
FIGURE 5.
Structural basis of SinR-DNA recognition. A, stereo view of the final 2Fobs − Fcalc electron density map colored in gray. Protein and DNA are represented by sticks, with atoms colored by type. The map is displayed at levels of 1.7 σ over the DNA and 1.2 σ over the protein so as not to obscure details in either component. B, stereo view of the SinR DNA binding domain in complex with double-stranded DNA containing a high affinity, inverted repeat SinR binding site taken directly from the eps promoter. The SinR α-helices are labeled, and DNA bases are drawn as colored squares as follows: A (red); T (blue); G (green), and C (yellow). C, expanded stereo view of the specific interactions by which SinR recognizes operator sequences. Contacts between protein side chains and DNA bases are shown as dotted lines with hydrogen bond donor or acceptor status indicated by lowercase lettering. It appears that SinR achieves its sequence specificity through a combination of direct and indirect readout; recognition of the first four bases of the 7-residue motif, 5′-GTTCTYT-3′, and the pyrimidine at position five is sufficient for sequence-specific binding. D, the sequence of the DNA used in structure determination; other than the 5′-A overhang, the sequence shown correlates perfectly to the region 72–53 bp upstream of the transcription start site of the eps promoter. E, schematic of the protein-DNA contacts in the crystal. DNA bases are all represented by hexagons, ribose sugars are represented by pentagons, and phosphates are represented by circles. Each DNA strand is colored separately, and the contacting amino acids are represented as white boxes.