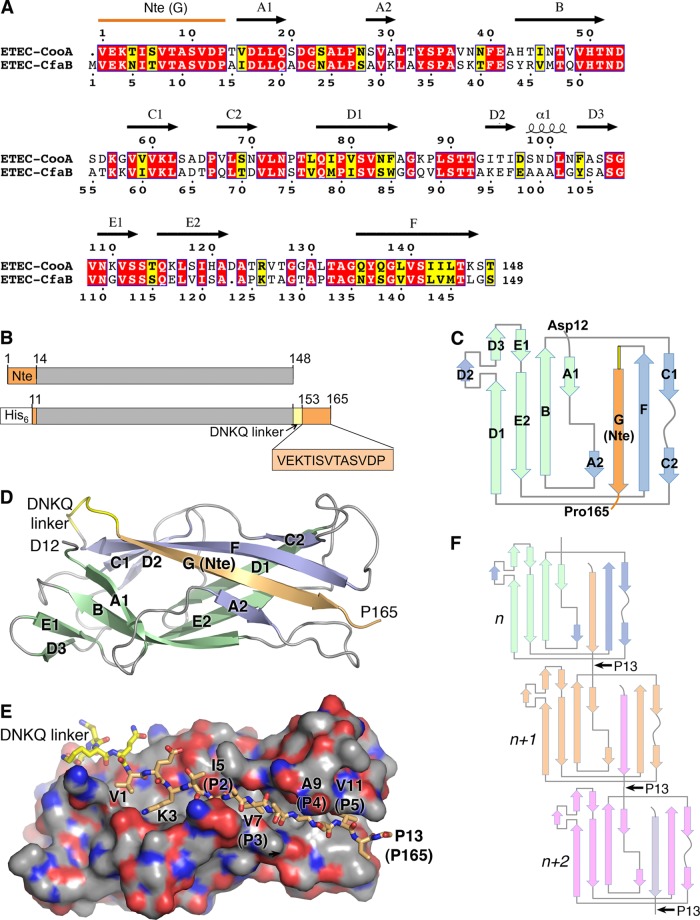
Fig 1.
Crystal structure of ETEC CooAdsc. (A) Amino acid sequence alignment of CooA and CfaB. Secondary structure elements of CooA are indicated above its sequence. Red background, identical residues; yellow background, similar residues. (B) Schematic representation of wild-type CooA (top) and the CooAdsc construct (bottom). The sequence shown with orange background represents the Nte, which has been removed from the N terminus and attached to the C terminus. (C) Schematic of the secondary structure of CooAdsc. Green β-strands belong to the first β-sheet and blue and orange strands belong to the second β-sheet in the Ig fold. The orange G strand is the Nte, which has been fused to the C terminus. (D) Ribbon representation of the 1.6-Å-resolution CooAdsc crystal structure. (E) Space-filling representation of CooA with the Nte shown in stick representation. Carbon atoms are colored orange in the Nte, yellow in the DNKQ linker, and gray in the remainder of the protein. Oxygen atoms are red, and nitrogens are blue. Residues in the Nte are numbered as in native mature CooA, where the Nte of subunit n+1 (not shown) is donated to subunit n (shown). The Nte fits into the hydrophobic groove with hydrophobic side chains at positions P2 to P5, indicated in parentheses, filling corresponding pockets in the groove. (F) CS1 assembly scheme showing donor strand complementation between adjacent CooA subunits. Each subunit has a different color to illustrate donor strand complementation with an adjacent subunit in the filament.