Abstract
Many pseudomonads produce redox active compounds called phenazines that function in a variety of biological processes. Phenazines are well known for their toxicity against non-phenazine-producing organisms, which allows them to serve as crucial biocontrol agents and virulence factors during infection. As for other secondary metabolites, conditions of nutritional stress or limitation stimulate the production of phenazines, but little is known of the molecular details underlying this phenomenon. Using a combination of microarray and metabolite analyses, we demonstrate that the assimilation of glycine as a carbon source and the biosynthesis of pyocyanin in Pseudomonas aeruginosa PAO1 are both dependent on the PA2449 gene. The inactivation of the PA2449 gene was found to influence the transcription of a core set of genes encoding a glycine cleavage system, serine hydroxymethyltransferase, and serine dehydratase. PA2449 also affected the transcription of several genes that are integral in cell signaling and pyocyanin biosynthesis in P. aeruginosa PAO1. This study sheds light on the unexpected relationship between the utilization of an unfavorable carbon source and the production of pyocyanin. PA2449 is conserved among pseudomonads and might be universally involved in the assimilation of glycine among this metabolically diverse group of bacteria.
INTRODUCTION
Phenazines are a large class of polyaromatic secondary metabolites produced by a variety of soil bacteria, particularly those belonging to the genera Streptomyces and Pseudomonas (1, 2). The high redox activity associated with phenazines enables them to function in diverse biological processes such as iron assimilation (3, 4), primary energy metabolism (5, 6), and cell-to-cell communication (7). Phenazines also serve as crucial antimicrobial agents, enabling bacteria to kill off competing microorganisms (8–10) and cause disease in a eukaryotic host during infection (11, 12). One of the better-known human pathogens that use phenazines as virulence factors is Pseudomonas aeruginosa (13, 14). Indeed, a hallmark feature of P. aeruginosa infections in individuals suffering from the genetic disorder cystic fibrosis is the production of the blue phenazine pyocyanin. Pyocyanin accumulates to such high levels (∼100 μM) that it gives the sputum of infected individuals a distinctly bluish hue (15).
There is an assortment of phenazine compounds biosynthesized by P. aeruginosa (16, 17), and all of them are derived from the common intermediate phenazine-1-carboxylic acid (PCA). In P. aeruginosa, PCA is biosynthesized from the central metabolite chorismate through a core group of seven enzymes known as PhzA, -B, -C, -D, -E, -F, and -G (18). PCA is then modified to produce more-complex phenazine derivatives by three tailoring enzymes, PhzH, PhzS, and PhzM (16). PhzH is an ATP-dependent aminotransferase that converts PCA into phenazine-1-carboxamide (PCN). The biosynthesis of pyocyanin consists of an S-adenoyslmethionine (SAM)-dependent N-methylation of PCA catalyzed by PhzM and then subsequent reduction of the carboxylic acid group via monooxygenase PhzS. PhzS can also oxidize PCA to give 1-hydroxyphenazine as an end product.
Interestingly, P. aeruginosa is one of the few human pathogens that generates phenazines via two independent, homologous pathways or gene clusters, phzA1B1C1D1E1F1G1-phzS and phzA2B2C2D2E2F2G2 (16). Whereas the phzS gene is in proximity to the phzA1 cluster, the phzM and phzH genes are localized elsewhere on the chromosome. Expression of the phzA1 locus accounts for the majority of phenazines produced by P. aeruginosa PAO1 (19). phzA1 expression is regulated by quorum-sensing (QS) cell signaling mechanisms (19, 20). The QS network in P. aeruginosa is composed of three distinct but overlapping systems: the two homoserine lactone (HSL) networks, N-3-oxo-dodecanoyl-HSL (3-oxo-C12-HSL) and N-butyryl-HSL (C4-HSL), and 2-akyl-4-quinolone (PQS)-mediated signaling. The autoinducer synthase and transcriptional regulator pairs LasI/LasR and RhlI/RhlR are responsible for the 3-oxo-C12-HSL and C4-HSL networks, respectively (21–23). Collectively, the 3-oxo-C12-HSL, C4-HSL, and PQS signaling systems cooperatively regulate the expression of numerous virulence factors, including exotoxins, proteases, rhamnolipids, and phenazines (21, 24).
The biosynthesis of phenazines in P. aeruginosa is orchestrated around an extensive network of environmental factors and protein regulatory elements. Classic environmental stresses known to induce phenazine production include phosphate limitation (25–27) and iron availability (28). Starvation of these essential nutrients has been found to positively impact QS, thus enhancing the expression of QS-related genes and phenotypes, including pyocyanin formation.
We have discovered that the metabolism of glycine as a carbon source may serve as an additional nutritional cue for the biosynthesis of pyocyanin in P. aeruginosa PAO1. The putative transcriptional regulator PA2449 gene in P. aeruginosa PAO1 was not only necessary for the production of pyocyanin but also essential for the assimilation of glycine as a carbon source. Understanding the relationship between glycine metabolism, pyocyanin biosynthesis, and the PA2449 gene was the main goal of the current study. Importantly, the results of our study provide a foundation for defining the role of PA2449 in the general metabolism and physiology of Pseudomonas-related bacteria.
MATERIALS AND METHODS
Media, growth conditions, and chemicals.
Bacteria were cultivated in BD Difco Lennox broth (LB) at 37°C unless otherwise stated. Solid bacteriological media were prepared with the addition of BD Bacto agar at 15 g liter−1. For the purposes of the experiments described herein, P. aeruginosa strains were grown in 50 ml of peptone broth (PB) in a 500-ml baffled shake flask at 200 rpm at 37°C. PB consisted of (per liter) 1.5 g MgSO4, 10 g K2SO4, and 20 g of BD BBL Gelysate peptone (29). Low-phosphate succinate (4), glycerol-alanine (30), nutrient yeast broth (31), and M9 and M63 minimal media (32) were prepared as described previously. M9 minimal medium was supplemented with 5 μM ferrous sulfate. Media were supplemented with kanamycin (50 μg ml−1), tetracycline (10 μg ml−1), and gentamicin (10 μg ml−1 for Escherichia coli and 30 μg ml−1 for P. aeruginosa) when needed for plasmid selection.
Chemicals were purchased from Fisher Scientific (Hampton, NJ) and used without further purification. Pyocyanin (catalog no. P0046) and N-(3-oxododecanoyl)-l-homoserine lactone (catalog no. O9139) were purchased from Sigma-Aldrich (St. Louis, MO). Purified phenazine-1-carboxylic acid was a kind gift from Linda Thomashow (USDA), and N-butyryl-l-homoserine lactone (catalog no. 10007898) was purchased from Cayman Chemical (Ann Arbor, MI).
Bacteria and plasmids.
The bacterial strains and plasmids used in this study are given in Tables S1 and S2 of the supplemental material, respectively. Plasmids were maintained in E. coli Top10 (Invitrogen, Carlsbad, CA) for cloning purposes. The wild-type P. aeruginosa PAO1-UW strain and its transposon mutants were acquired from the P. aeruginosa PAO1 transposon mutant library (33). All P. aeruginosa PAO1-UW mutants were verified for transposon insertions at proper loci through PCR as recommended by the library curators (University of Washington Department of Genome Sciences). Plasmids were introduced into P. aeruginosa by electroporation using established methods (34). Recombinant P. aeruginosa strains were selected on LB supplemented with 30 μg ml−1 of gentamicin at 37°C for 18 h.
Molecular biology methods.
DNA was purified using Promega (Madison, WI) nucleic acid purification kits. Restriction enzymes and ligases were products of New England BioLabs (Ipswich, MA). PrimeStar polymerase (TaKaRa Biosciences, Japan) was used for all PCRs. PCR applications were done by following the recommended protocols for the PrimeStar polymerase and with the oligonucleotides given in Table S3 in the supplemental material. Genomic DNA from P. aeruginosa PAO1-UW was used for all PCR applications. All desired PCR products were gel purified and cloned into pCR-Blunt according to the manufacturer's instructions (Invitrogen, Carlsbad, CA). Cloned DNA was verified by sequencing (Genewiz, South Plainfield, NJ).
Construction of plasmids pBRL413, pBRL435, and pBRL480.
The DNA region containing the putative PA2449 open reading frame (ORF) was amplified by PCR. The gel-purified PA2449 PCR product was cloned into pCR-Blunt to give the plasmid pBRL408. The PA2449 ORF was subcloned from pBRL408 into the NdeI/EcoR I restriction sites of pET28b (Novagen) to yield the plasmid pBRL417. Subsequently, pBRL417 was digested with XbaI and SacI to liberate the PA2449 ORF with a 5′ ribosome binding site (RBS), which was then ligated into the XbaI/SacI sites of pBBR1MCS-5 to generate the plasmid pBRL435. Plasmid pBRL413 was created by subcloning the HindIII phzA2B2C2D2E2F2 fragment from pUCP-A2G2 (16) into the corresponding HindIII site of pBBR1MCS-5.
The metE gene was PCR amplified from genomic DNA of E. coli K-12 MG1655. The desired metE product was gel purified and cloned into pCR-Blunt to give plasmid pBRL409. The metE gene was subcloned from pBRL409 into the KpnI/SacI sites of pBBR1MCS-5 to yield plasmid pBRL480.
Construction of phzB1, phzB2, phzM, and phzH-lacZ reporter plasmids.
We first constructed a template plasmid for measuring promoter activity in P. aeruginosa based on lacZ expression. First, the lacZ ORF was PCR amplified from E. coli K-12 MG1655 genomic DNA with the primers BL393.f and BL393.r (see Table S3 in the supplemental material). The BL393.f primer installed a pET-derived RBS at the 5′ end of the lacZ ORF. The gel-purified RBS-lacZ product was cloned into pCR-Blunt to give pBRL410. The RBS-lacZ fragment was subsequently cloned from pBRL410 into the KpnI/HindIII sites of pBBR1MCS-5 to yield pBRL416. Promoters could then be inserted upstream of the RBS-lacZ region via directional cloning with 5′ SphI and 3′ KpnI restriction sites.
Promoter regions for phzB1, phzB2, phzM, and phzH were amplified by PCR. The desired PCR-amplified promoter regions were gel purified and cloned into pCR-Blunt (see Table S2 in the supplemental material). Subsequently, the promoter regions of phzB1, phzB2, phzM, and phzH were individually subcloned into the SphI/KpnI restriction sites of pBRL416 to give pBRL418, pBRL423, pBRL424, and pBRL425, respectively.
Construction of gcvP2, hcnA, and metE lacZ reporter plasmids.
We found that expression of lacZ from the phz promoters was slightly higher than desired (Miller units were >10,000). This was attributed to the pET-derived RBS. Although this was not a significant problem, we circumvented this concern for the gcvP2, hcnA, and metE promoter-lacZ reporter constructs by directly fusing the 5′ promoter region with the E. coli lacZ ORF. Thus, lacZ translation in these constructs was dictated by the native RBS of the gcvP2, hcnA, or metE promoter region. First, the lacZ ORF was PCR amplified from E. coli K-12 MG1655 genomic DNA, and the desired lacZ product was gel purified. Next, the promoter regions of gcvP2, hcnA, and metE were amplified by PCR with primers that installed a 3′ end homologous region into the E. coli lacZ ORF (see Table S3 in the supplemental material). Following gel purification, the gcvP2, hcnA, and metE promoters were individually joined with the E. coli lacZ ORF using fusion PCR (35). The desired promoter-lacZ fusion constructs were gel purified and subsequently cloned into pCR-Blunt (see Table S2 in the supplemental material). The gcvP2-, hcnA-, and metE-lacZ constructs were subcloned into the KpnI/SphI, KpnI/SphI, and HindIII/SphI restriction sites of pBBR1MCS-5 to give the plasmids pBRL456, pBRL447, and pBRL415, respectively.
Measurement of galactosidase activity.
The activity of β-galactosidase was determined for recombinant P. aeruginosa PAO1 strains using the Miller assay (36).
Staphylococcus aureus killing assays.
In triplicate, P. aeruginosa PAO1 and PW5126 were grown in PB to an optical density at 600 nm (OD600) of 0.7. At this time, 2 μl of each culture was spotted onto an LB plate that was freshly swabbed with LB-grown (37°C, 200 rpm, 24 h) Staphylococcus aureus ATCC 25923. Spotted plates were grown at 37°C for 24 h.
Spectrophotometric measurement of extracellular pyocyanin.
Pyocyanin was measured using reported methods (7). Briefly, cells were removed from cultures by centrifugation (16,000 × g for 5 min) and subsequent passage through Acrodisc syringe filters with 0.2-μm nylon membranes. The absorbances of the cell-free samples at 690 nm were measured and converted into pyocyanin concentrations using a molar extinction coefficient of 4,130 M−1 cm−1 (37).
Growth of P. aeruginosa with amino acids serving as the sole carbon or nitrogen source.
Analysis of the growth of P. aeruginosa with amino acids serving as the sole carbon or nitrogen source was done in triplicate. Both P. aeruginosa PAO1 and PW5126 were initially grown on LB at 37°C for 18 h. Individual clones were then patched onto solid minimal salt media (50 mM Na2HPO4, 20 mM KH2PO4, 10 mM NaCl, 2 mM MgSO4, 0.1 mM CaCl2, 5 μM FeSO4, pH 7.0), which was supplemented with either 20 mM (carbon source) or 5 mM (nitrogen source) l-alanine, l-arginine, l-asparagine, l-aspartate, l-glutamate, l-histidine, l-isoleucine, glycine, l-leucine, l-lysine, l-phenylalanine, proline, l-tryptophan, l-tyrosine, l-serine, and l-valine. When testing amino acids as carbon or nitrogen sources, the minimal media were further supplemented with 20 mM NH4Cl or 20 mM succinate, respectively. Patched plates were incubated at 37°C for 1 to 5 days.
For liquid cultures, minimal salt medium (given above) was supplemented with either glycine, l-valine, l-serine, or l-glutamine to a final concentration of 20 mM (carbon source) or 10 mM (nitrogen source). This medium was further supplemented with 20 mM NH4Cl or 20 mM succinate depending on whether the amino acid was supplied as a carbon or nitrogen source, respectively. Strains were inoculated into 50 ml of minimal media to an initial OD600 of ∼0.1. At 12, 24, 36, 48, 60, 72, and 84 h postinoculation, 2.5 ml of minimal salt medium (does not have a source of carbon or nitrogen) was added to each culture. This addition compensated for volume loss due to both sampling and evaporation. Cultures were grown at 37°C, 200 rpm for 12 to 96 h.
ESI-LC-MS analysis of phenazines.
In triplicate, P. aeruginosa PAO1 and PW5126 strains were grown in 50 ml PB at 37°C and 200 rpm for 24 h. Cells were removed via centrifugation (5,000 × g for 20 min), and the resulting supernatants were extracted with an equal volume (50 ml) of chloroform. Chloroform extracts were evaporated to dryness. The dried-phenazine samples were resuspended in 500 μl acetonitrile and vortexed vigorously for 5 min, and the insoluble material was removed by centrifugation at 5,000 × g for 5 min. The resulting supernatants (1 μl) were used for electrospray ionization-liquid chromatography-mass spectrometry (ESI-LC-MS).
ESI-LC-MS analysis was performed on an API2000 LC/tandem MS (LC/MS/MS) system equipped with a TurboIon Spray source interfaced with a Prominence ultrafast liquid chromatograph (UFLC). A reverse-phase BDS Hypersil C18 column (100 mm by 2.1-mm inside diameter [ID], 3-μm particle size,) was employed. Mobile phases A (5% acetonitrile:95% water with 0.05% formic acid) and B (95% acetonitrile:5% water with 0.05% formic acid) were used for all experiments. Phenazine separation was performed using a gradient program of 3 min of 100% A, a 40-min linear gradient to 100% B, and 10 min of 100% B at a constant flow rate of 250 μl min−1. Electrospray ionization in the positive mode was performed using the TurboIon Spray source with an ion spray voltage of 5,500 V, a desolvation temperature of 250°C, and gas 1 and gas 2 set at 20 and 30, respectively. Mass spectra were collected over the m/z range of 150 to 300 with a declustering potential of 22 V, a focusing potential of 400 V, and an entrance potential of 5 V.
ESI-LC-MS analysis of HSLs.
In duplicate, P. aeruginosa PAO1 and PW5126 were grown in 100 ml of PB at 37°C and 200 rpm to an OD600 of ∼0.6. Cells were then removed by centrifugation (5,000 × g for 20 min), and the resulting supernatants were then extracted three successive times with equal volumes (100 ml) of dichloromethane (DCM). The three 100-ml DCM fractions were combined and evaporated to dryness. The dried residues containing the extracted HSLs were suspended in 500 μl of acetonitrile and then centrifuged at 16,000 × g for 10 min to remove insoluble material. The resulting supernatants (1 μl) were used for ESI-LC-MS. For quantification of HSLs present in the samples, standard curves were generated for both authentic C4-HSL and 3-oxo-C12-HSL. Lower limits of detection for C4-HSL and 3-oxo-C12-HSL were determined to be 50 pmol and 10 pmol, respectively.
ESI-LC-MS analysis was performed using the same instrumentation and column described above. Mobile phases A (water with 0.05% formic acid) and B (90% methanol:10% water with 0.05% formic acid) were employed. HSL separation was done using an isocratic profile of 10% phase B in phase A for 1 min, a 7-min linear gradient to 100% B, and 4 min of 100% B at a constant flow rate of 0.25 μl min−1. Electrospray ionization in the positive mode was performed using the TurboIon Spray source with an ion spray voltage of 4,500 V, a desolvation temperature of 300°C, and gas 1 and gas 2 set at 20 and 30, respectively. Mass spectra were collected over the m/z range of 100 to 300 with a declustering potential of 22 V, a focusing potential of 400 V, and an entrance potential of 5 V.
HPLC analysis of free amino acids.
In triplicate, P. aeruginosa PAO1 and PW5126 were grown in 50 ml PB at 37°C and 200 rpm for 24 h. At 6 and 24 h postinoculation, 5 ml of culture was removed and cleared of cells via centrifugation at 5,000 × g for 20 min. The cell-free supernatants were passed through an Amicon 5,000-molecular-weight-cutoff (MWCO) centrifugal filter. The filter was washed with 5 ml of H2O, and the collected filtrates (10 ml) were lyophilized. Uncultured PB (control) was processed in an identical manner for amino acid analysis. The freeze-dried samples were resuspended in 2 ml of high-pressure LC (HPLC) grade H2O, vigorously vortexed for 5 min, and then incubated at 37°C for 30 min. Insoluble material was removed by centrifugation at 5,000 × g for 5 min.
HPLC was performed using an 1100 series HPLC-diode array detector (HPLC-DAD) system (Agilent Technologies, Santa Clara, CA). The system consisted of a programmable autosampler with a 100-μl built-in loop, a quaternary pump, a column thermostat, a photodiode array detector, and Chemstation software (version B.03.02). Amino acids were derivatized using an automated online method and subsequently separated on a reverse-phase Agilent Zorbax Eclipse amino acid analysis (AAA) column (150 mm by 4.6-mm ID, 3.5-μm particle size) maintained at a temperature of 40°C. Reagents used for derivatization were purchased from Agilent, including o-phthaldehyde (OPA) for primary amino acids and 9-fluorenylmethyl chloroformate (FMOC) for secondary amino acids. Detailed information for the setup and execution of this method has been published (38). Chemical separations employed mobile phases A (40 mM Na2HPO3, pH 7.8) and B (45% acetonitrile:45% methanol:10% H2O) in a gradient program consisting of 1.9 min of 100% phase A, a 16.2-min linear gradient to 57% phase B, 4.1 min of 100% phase B, and 2.1 min of 100% phase A at a flow rate of 2 ml min−1. UV detection used a setting of 338 nm, 10 nm bandwidth. Standard curves of glycine and serine were constructed to measure the concentrations of glycine and serine in each individual sample.
cDNA microarray analysis.
In quadruplicate, P. aeruginosa PAO1 and PW5126 were grown at 37°C in 50 ml of PB in 500-ml baffled shake flasks at 200 rpm to an OD600 of 0.7. At this time, 0.5 ml of culture was immediately mixed with 1 ml of RNAprotect bacterial reagent (Qiagen, Valencia, CA). The bacteria in the treated samples were lysed using an enzymatic proteinase K digestion approach as detailed in the RNAprotect bacterial reagent handbook (Qiagen). The RNA was immediately purified using the RNeasy minikit (Qiagen) with an on-column DNase digestion step to remove contaminating DNA. Purified RNA samples were checked for DNA contamination by PCR with the primers BL343.f and BL343.r (see Table S3 in the supplemental material), which were designed to amplify the rplU gene (39). Purified RNA samples were also analyzed for quality using a Bioanalyzer (Agilent).
Microarray studies were carried out by the Microarray Core Facility (Upstate Medical University, Syracuse, NY) using GeneChip P. aeruginosa PAO1 Affymetrix arrays. Experiments were performed according to the Affymetrix GeneChip expression analysis technical manual (Affymetrix publication 702232, revision 3) and methods established at the State University of New York (SUNY) Upstate (Syracuse, NY). For initial data processing, the Affymetrix software (MicroArray suite 5.0 [MAS5]) was used for quality control (Q/C) analysis, which involved calculating the signal intensities from each perfect match proved on the arrays relative to those for the mismatch probe and determining whether or not the gene was present in the sample. The robust multiarray average (RMA) method was used to normalize the set of arrays in our screen (GeneTraffic software; Stratagene, La Jolla, CA). The MultiExperiment viewer (MeV; v4.6.2) was used for subsequent statistical analysis. To identify genes displaying a significant difference in intensities, a t test between subjects was performed using P values based on all permutations and an overall alpha value fixed at 0.05. False discoveries were addressed using the standard Bonferroni correction.
Microarray database accession number.
Data from the microarray experiments, including all CEL files, were deposited in Gene Expression Omnibus (GEO) database with the accession number GSE39044.
RESULTS
The PA2449 ORF is predicted to encode a transcriptional regulator belonging to the TyrR family.
In addition to being a putative TyrR-like protein, the PA2449 protein also possesses homology to transcriptional regulators known as enhancer binding proteins (EBPs) that interact with the alternative sigma factor σ54 (RpoN) to activate transcription (40). The RpoN interaction domain is conserved in the PA2449 protein(see Fig. S1 in the supplemental material). In P. aeruginosa PAO1 there exists another TyrR-like EBP transcriptional regulator known as PhhR (41). Previous studies found that PhhR functions as an EBP for the transcriptional activation of genes involved in tyrosine and phenylalanine catabolism (42), which consequently enhanced the production of pyocyanin (43). Because the PA2449 protein displays homology to both PhhR (44% identity) of P. aeruginosa and TyrR (41% identity) of E. coli, we suspected that PA2449 might also be involved in phenazine biosynthesis.
Disruption of the PA2449 gene eliminates pyocyanin production.
A P. aeruginosa PAO1 strain possessing a transposon insertion within the PA2449 gene was obtained from the P. aeruginosa PAO1 transposon mutant library (33). This strain, designated PW5126, was found to be incapable of producing measurable quantities of the blue pigment pyocyanin in a variety of rich and minimal media (Fig. 1). For example, PW5126 did not produce pyocyanin when grown in a common rich medium such as Lennox, Luria-Bertani, or nutrient yeast broth. In addition, pyocyanin was not detected when PW5126 was cultivated in M9 and M63 minimal media supplemented with various carbon sources (20 mM), including glucose, glycerol, succinate, and citrate. Low-phosphate succinate medium (LPSM) and glycerol-alanine (GA) medium are two types of minimal media that stimulate the formation of high levels of pyocyanin (4, 30). However, PW5126 did not generate detectable levels of pyocyanin when grown in either LPSM or GA medium.
Fig 1.
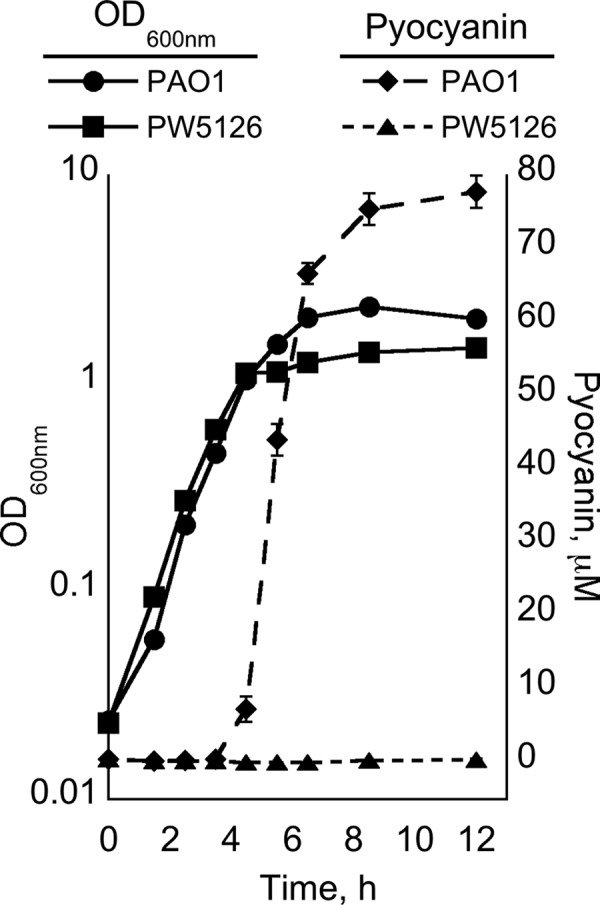
Pyocyanin production from P. aeruginosa PAO1 and PW5126 grown in PB. Cultures were assayed for OD600 and pyocyanin via spectrophotometry. Individual points represent mean values of triplicate samples, and standard error bars are shown.
Peptone broth (PB) is an established medium for enriching the production of pyocyanin from P. aeruginosa (29). PB is the liquid version of Pseudomonas isolation agar (PIA), a well-known and classic solid medium used for identifying P. aeruginosa based on pyocyanin formation (44). A key formulation of PB that contributes to enhancing pyocyanin production is the use of a peptone containing a high percentage (∼20%) of glycine and limiting amounts (<0.2 mM) of phosphate. Either bacteriological or gelatin-based peptones meet these criteria. As with other media tested in our study, PW5126 did not produce detectable quantities of pyocyanin when cultured in gelatin-based PB (Fig. 1 and 2). Therefore, because of the historical importance of PIA, we focused our experiments on defining the relationship between the PA2449 gene and pyocyanin biosynthesis in P. aeruginosa PAO1 when grown in gelatin-based PB.
Fig 2.
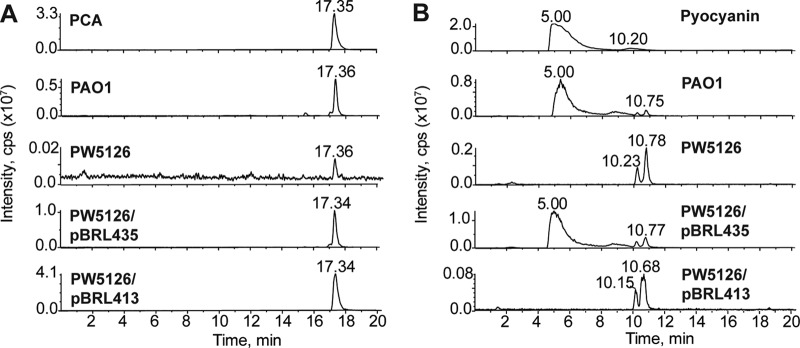
LC-MS analysis of phenazines produced from P. aeruginosa PAO1 and PW5126. Strains were grown in PB for 24 h at 37°C. Total phenazines were extracted and subsequently resuspended in water-methanol for LC-MS. Ion extraction of m/z 224.75 to 225.5 and m/z 210.75 to 211.5 was used to detect the [M+H]+ for PCA (A) and pyocyanin (B), respectively. In contrast to P. aeruginosa PAO1, PW5126 did not produce pyocyanin but generated trace quantities of PCA. Expression of the PA2449 ORF from the lac promoter of pBBR1MCS-5 (pBRL435) restored PCA and pyocyanin biosynthesis in PW5126. Expression of the phzA2B2C2D2E2F2G2 cluster from the lac promoter of pBBR1MCS-5 (pBRL413) allowed PW5126 to produce PCA but not pyocyanin. These data indicate that production of pyocyanin in P. aeruginosa PAO1 requires the PA2449 gene.
PA2449 is required for pyocyanin production in PB.
To verify that the pyocyanin deficiency of PW5126 was the result of the transposon insertion in the PA2449 gene, the PA2449 ORF of P. aeruginosa PAO1 was cloned into the broad-host-range vector pBBR1MCS-5 and the resulting plasmid (pBRL435) was electroporated into PW5126. The recombinant PW5126/pBRL435 strain successfully biosynthesized pyocyanin (Fig. 2), although the levels were ∼65% less than those of P. aeruginosa PAO1 harboring pBBR1MCS-5. The reduced pyocyanin concentrations could have been due to low expression associated with the lac promoter of the pBBR1MCS-5 parent vector. Nonetheless, the sole expression of the PA2449 ORF was sufficient to restore pyocyanin production in PW5126, thus confirming that a functional PA2449 gene is required for the biosynthesis of pyocyanin in P. aeruginosa PAO1.
We next determined the overall population and relative amounts of phenazines biosynthesized by PW5126 after 24 h of growth in PB. Total phenazines were extracted from either P. aeruginosa PAO1 or PW5126 cultures using chloroform, and the resulting chloroform extracts were analyzed for individual phenazine compounds by LC-MS (Fig. 2). The only phenazine derivative successfully produced by PW5126 was PCA at trace amounts (∼0.5% of wild-type levels). This indicated that PCA biosynthesis was still operative in the absence of PA2449, albeit at extremely low levels.
It was initially believed that the absence of the PA2449 gene could disrupt the biosynthesis of key metabolites, e.g., chorismate, that are necessary for the production of phenazines. However, PW5126 readily synthesized PCA when the phzA2B2C2D2E2F2G2 gene cluster was heterologously expressed from a lac promoter (Fig. 2), suggesting that precursor availability was not a concern. We next examined if the PA2449 gene was necessary for activating or upregulating the expression of genes required for phenazine biosynthesis. This was accomplished by measuring the expression of the phz-related genes (phzB1, phzB2, phzM, and phzH) using β-galactosidase (lacZ) reporter assays (Fig. 3). For P. aeruginosa PAO1 cultures, there was a significant change in the expression of the phzB1-, phzB2-, and phzM-lacZ constructs, which began approximately 1 h prior to the visible accumulation of pyocyanin in PB. This change in phzB1, phzB2, and phzM expression was not observed for the transposon insertion PA2449 mutant. For PW5126, there was a >95% reduction in phzB1-lacZ expression (Fig. 3A), whereas phzB2- and phzM-lacZ expression decreased by ∼60 and 80%, respectively. Expression of the phzH-lacZ construct was not affected by the PA2449 mutation (Fig. 3D). These results revealed that the probable cause for the pyocyanin-deficient phenotype of PW5126 was insufficient expression of the phz genes that are necessary for the biosynthesis of phenazines in P. aeruginosa PAO1.
Fig 3.
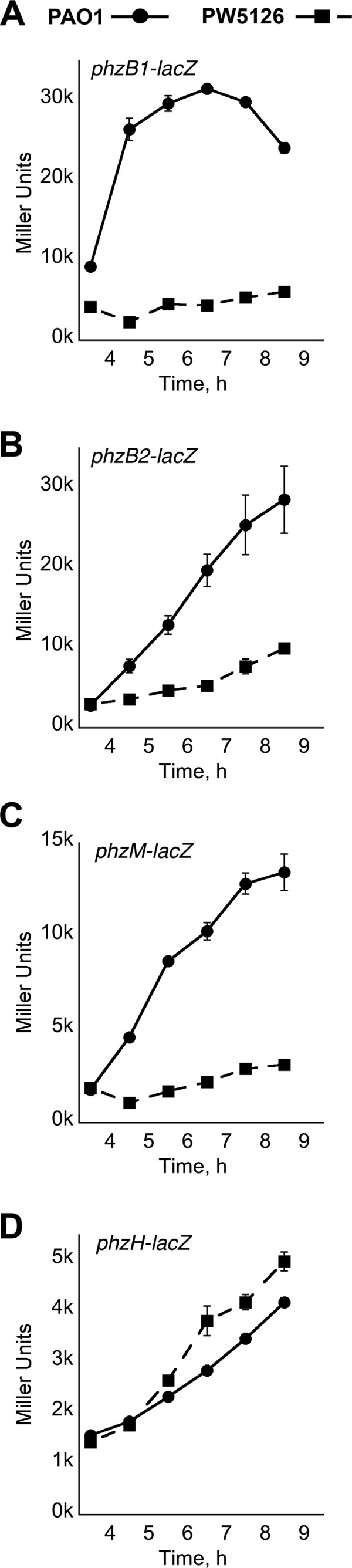
Relative expression levels of the phzB1, phzB2, phzM, and phzH genes in P. aeruginosa PAO1 and PW5126. The 5′ regulatory DNA regions consisting of either ∼1,000 bp (phzB1 [A], phzB2 [B], and phzM [C]) or 500 bp (phzH [D]) of the designated phz ORFs were cloned upstream of an E. coli lacZ ORF in a promoter-less pBBR1MCS-5 plasmid. P. aeruginosa strains harboring the phz-lacZ constructs were grown in PB at 37°C and periodically assayed for LacZ activity. Individual points represent mean values of triplicate samples, and standard error bars are shown.
PA2449 is necessary for the production of C4-HSL in PB.
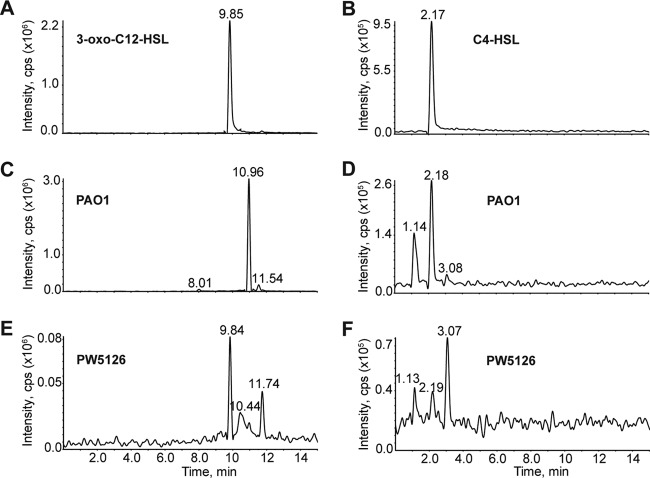
The signaling molecule C4-HSL and its cognate receptor, RhlR, positively regulate the expression of the phzA1 locus (19). Our concern was therefore that the pyocyanin-deficient phenotype of PW5126 was due to low or insufficient levels of HSLs, e.g., C4-HSL, as a result of the PA2449 mutation. Low levels of HSLs would fail to fully activate expression of the phz genes as observed for PW5126 (Fig. 3), so we decided to assay for HSL production by LC-MS (45). Both P. aeruginosa PAO1 and PW5126 were grown in PB to an OD600 of ∼0.7. This cell density represented a crucial point for the analysis of QS in its relation to pyocyanin production because (i) P. aeruginosa PAO1 cultures just began to display a distinctive bluish hue indicative of pyocyanin and (ii) it was 1 h after transcriptional activation of the phz genes as determined by lacZ reporter assays (Fig. 3). Both sets of cultures were cleared of cellular material and subsequently extracted with dichloromethane to isolate total HSLs. The HSL extracts were analyzed by LC-MS for 3-oxo-C12-HSL and C4-HSL (Fig. 4). Under pyocyanin-producing conditions in PB, P. aeruginosa PAO1 produced only C4-HSL (0.3 ± 0.06 μM) and no detectable quantities of 3-oxo-C12-HSL. In contrast, PW5126 did not produce measurable levels of C4-HSL but small amounts (0.07 ± 0.03 μM) of 3-oxo-C12-HSL were observed. The concentrations of C4-HSL produced from PW5126 cells, compared to those produced from P. aeruginosa PAO1, were not sufficient to activate the expression of the phz loci needed for the biosynthesis of pyocyanin (Fig. 3). Additionally, insufficient levels of C4-HSL and subsequent loss of transcriptional activation of phz genes could account for the residual levels of PCA observed from PW5126 cultures (Fig. 2). However, the exogenous addition of HSLs to PW5126 cultures did not restore pyocyanin production, thereby suggesting that involvement of PA2449 is more complex than simply via HSL mechanisms.
Fig 4.
LC-MS analysis of HSLs produced from P. aeruginosa PAO1 and PW5126. Strains were grown in PB at 37°C to an OD600 of 0.7. Total HSLs were extracted and subsequently resuspended in acetonitrile for LC-MS analysis. HSLs were detected by ion extraction of their respective [M+H]+ m/z's. Chromatograms are shown for authentic 3-oxo-C12-HSL (m/z of 298 to 299) (A) and C4-HSL (m/z of 172 to 173) (B). Wild-type P. aeruginosa PAO1 did not produce detectable levels of 3-oxo-C12-HSL (C), but C4-HSL (D) was observed. Conversely, PW5126 produced 3-oxo-C12-HSL (E) and insignificant quantities of C4-HSL (F).
For P. aeruginosa, 2-alkyl-4-quinolones (PQS) act as signaling molecules and have been shown to facilitate the expression of rhlI and rhlR required for C4-HSL biosynthesis (46). If the production of PQS compounds was deficient in PW5126, then this would translate into low levels of C4-HSL. In addition to their role as signaling molecules in P. aeruginosa, PQS and related 4-quinolone compounds are necessary for the killing of several Gram-positive bacteria, including Staphylococcus aureus, by P. aeruginosa (47–49). We therefore examined (qualitatively) if PW5126 was deficient in the production of quinolones by determining its ability to lyse S. aureus. Both P. aeruginosa PAO1 and PW5126 were grown in PB to an OD600 of ∼0.7 and subsequently spotted onto LB plates that had been freshly swabbed with S. aureus. After 24 h of incubation at 37°C, we observed that PW5126 produced the same relative level of S. aureus killing as P. aeruginosa PAO1 (see Fig. S2 in the supplemental material). The inactivation of the PA2449 gene did not appear to have an overall negative effect on quinolone biosynthesis. A more detailed analysis of the spatial and temporal distribution of individual PQS signaling molecules is required to gain an accurate picture on how the disruption of the PA2449 gene affects the PQS network in P. aeruginosa PAO1.
Use of glycine as a sole carbon source requires PA2449.
As shown in Fig. 1, P. aeruginosa PAO1 and PW5126 had similar growth profiles in PB. However, PW5126 displayed a distinct cessation of growth at an OD600 of ∼1.0, which lasted for approximately 1 h until growth resumed at a slower pace than that of P. aeruginosa PAO1. Notably, the cessation of growth of PW5126 occurred slightly after the onset of pyocyanin accumulation in P. aeruginosa PAO1 cultures. Because amino acids serve as the only carbon sources in PB (50), we believed that an inability to utilize particular amino acids might be hindering the growth of PW5126. To test this hypothesis, PW5126 was grown on minimal agar plates supplemented with individual amino acids as either the sole carbon or nitrogen source. Glycine, serine, and valine were the only amino acid carbon sources that did not support colony formation of PW5126 even following a prolonged incubation of 5 days at 37°C. Subsequent analysis in liquid culture revealed that both serine and valine were suitable carbon sources for PW5126 (see Fig. S3 in the supplemental material), although overall cell yields on either amino acid were significantly lower (>60%) than those observed for P. aeruginosa PAO1. For glycine-grown PW5126, the final cell densities were ∼90% lower than those of P. aeruginosa PAO1 and did not increase from their initial values for the duration (96 h) of the experiment (see Fig. S3 in the supplemental material). The assimilation of glycine as a carbon source was dependent on the PA2449 gene in P. aeruginosa PAO1.
All tested individual amino acids were acceptable nitrogen sources for PW5126 and enabled the formation of colonies within 24 h, with succinate serving as the carbon source. The only exception to these findings was glycine, in which PW5126 took twice as long (48 h) to form visible colonies as P. aeruginosa PAO1. This observation was confirmed in liquid culture, where there was a >50% reduction in cell yields of PW5126 when glycine was the nitrogen source (see Fig. S3 in the supplemental material). In conclusion, the PA2449 gene was essential for the growth of P. aeruginosa PAO1 on glycine as a sole carbon but not a nitrogen source.
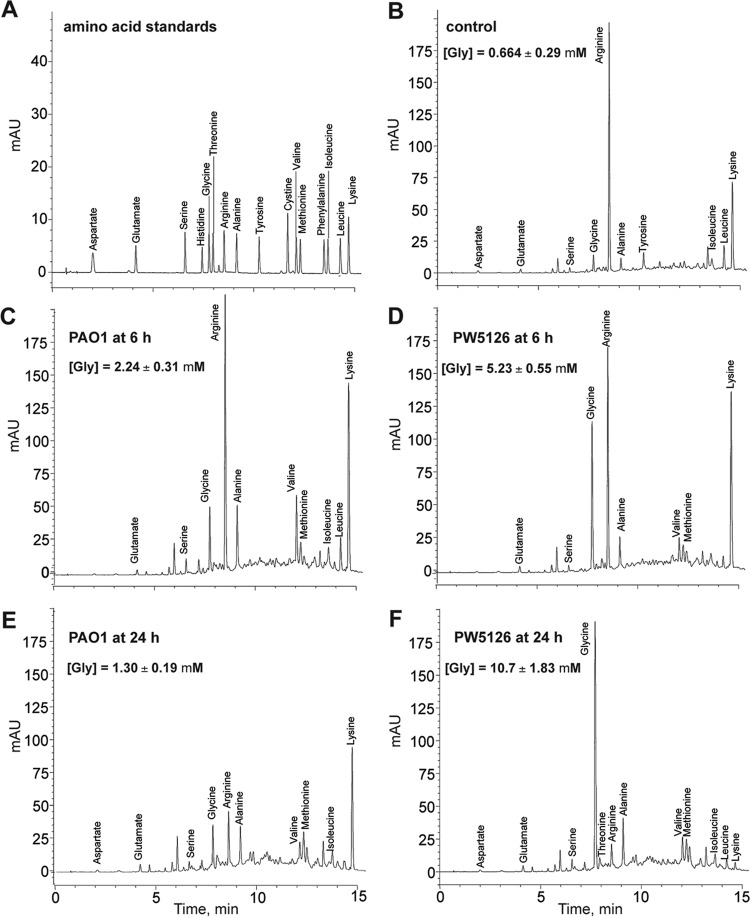
As stated earlier, the peptone used in PB is composed of ∼20% glycine bound in the form of oligopeptides, whereas free glycine is <0.5% (50). These percentages give a total glycine concentration of ∼40 mM (free glycine is <1 mM) for the quantity of peptone used in PB medium. The growth arrest observed for PW5126 when cultured in PB might therefore be due to an inadequacy in the assimilation of glycine as it is released from the oligopeptides through the actions of intrinsic proteases/peptidases. To assay for glycine assimilation, spent PB media derived from either P. aeruginosa PAO1 or PW5126 cultures were analyzed for free amino acid content at 6 and 24 h postinoculation by HPLC. Overall, the spent PB samples from both sets of cultures displayed similar trends in amino acid composition except for glycine (Fig. 5). Whereas the measured extracellular free glycine remained relatively constant at ∼1 mM in P. aeruginosa PAO1 cultures, we observed a continuous increase or buildup of free glycine from ∼1 to 10 mM for PW5126. The predominant remaining amino acid for PW5126 at 24 h was glycine. The elevated glycine concentrations generated in PW5126 cultures are indicative of a deficiency in glycine assimilation and are consistent with PA2449 being involved in glycine metabolism for P. aeruginosa PAO1.
Fig 5.
HPLC analysis of free amino acids present in spent PB media of P. aeruginosa PAO1 and PW5126. Strains were grown in PB at 37°C. At 6 and 24 h postinoculation, free amino acids present in spent media were derivatized and subsequently separated on a Zorbax Eclipse AAA column. Represented chromatograms are shown for an amino acid mixture (A), uninoculated PB (B), PAO1 at 6 h (C), PW5126 at 6 h (D), PAO1 at 24 h (E), and PW5126 at 24 h (F). Glycine concentrations represent mean values of triplicate samples, and standard deviations are shown.
Inactivation of the PA2449 gene affects the transcription of genes involved in glycine metabolism and cell signaling.
PA2449 was observed to be essential for both pyocyanin biosynthesis and the utilization of glycine, and its deletion had a deleterious effect on the synthesis of C4-HSL. To elucidate how these processes are related through the PA2449 gene, we used microarrays to determine the transcriptome of PW5126. It was expected that the resulting transcriptomic data would provide key information on the involvement of PA2449 in both glycine assimilation and the biosynthesis of pyocyanin. In quadruplicate, P. aeruginosa PAO1 or PW5126 was grown in PB to an OD600 of ∼0.7, at which time total RNA was isolated and subsequently processed for analysis with Affymetrix GeneChip P. aeruginosa microarrays. The microarray results identified a total of ∼300 genes that were statistically different in transcript expression by >2-fold between PW5126 and P. aeruginosa PAO1 (see Table S4 in the supplemental material).
As established earlier, disruption of the PA2449 gene significantly reduced the production of C4-HSL in PB. Expectedly, the majority of genes found to be transcriptionally downregulated in PW5126 were previously reported to under the regulation of C4-HSL in P. aeruginosa PAO1 (Table 1). The most notable members of this group were genes required for phenazine biosynthesis: phzA1B1C1D1E1F1G1, phzA2B2C2D2E2F2G2, phzS, and phzM. The phz loci were upregulated by >30-fold in P. aeruginosa PAO1 compared to PW5126. The failure to activate expression of the phz loci, which is supported from both microarray and lacZ reporter experiments, would account for the inability of PW5126 to produce pyocyanin. The microarray data also provided some insight into how the disruption of PA2449 may exert its influence on C4-HSL biosynthesis. The rhlI gene, encoding the C4-HSL synthase, was downregulated 3-fold in PW5126. The rhlI gene was the only QS regulatory protein gene downregulated; the lasI, lasR, rhlR, and PQS-related genes displayed no differences in transcript levels.
Table 1.
Selected list of genes having >2-fold changes in transcript levels in a transposon insertion PA2449 mutant (PW5126) compared to wild-type P. aeruginosa PAO1 grown in PB
| Function and gene name(s) | Gene IDa | Mean fold change ± SD | Biological function of product |
|---|---|---|---|
| Phenazine biosynthesis | |||
| phzA1, phzA2 | PA1899, PA4210 | −24.3 ± 4.69 | Biosynthesis of PCA core |
| phzB1, phzB2 | PA1900, PA4211 | −113 ± 16.8 | Biosynthesis of PCA core |
| phzC1, phzC2 | PA1901, PA4212 | −30.7 ± 9.04 | Biosynthesis of PCA core |
| phzD1, phzD2 | PA1902, PA4213 | −27.5 ± 4.87 | Biosynthesis of PCA core |
| phzE1, phzE2 | PA1903, PA4214 | −45.7 ± 9.59 | Biosynthesis of PCA core |
| phzF1, phzF2 | PA1904, PA4215 | −46.8 ± 12.3 | Biosynthesis of PCA core |
| phzG1, phzG2 | PA1905, PA4216 | −58.6 ± 7.83 | Biosynthesis of PCA core |
| phzS | PA4217 | −68.9 ± 8.75 | Monooxygenase |
| phzM | PA4209 | −22.7 ± 2.79 | Methyltransferase |
| Glycine metabolism | |||
| hcnA | PA2193 | −38.2 ± 5.10 | HCN synthase |
| hcnB | PA2194 | −11.9 ± 4.02 | HCN synthase |
| hcnC | PA2195 | −9.10 ± 2.19 | HCN synthase |
| gcvT2 | PA2442 | −12.6 ± 2.34 | GCSb protein T |
| sdaA | PA2443 | −11.7 ± 1.32 | Serine dehydratase |
| glyA2 | PA2444 | −54.4 ± 6.04 | Serine hydroxymethyltransferase |
| gcvP2 | PA2445 | −37.5 ± 4.58 | GCS protein P |
| gcvH2 | PA2446 | −59.3 ± 25.2 | GCS protein H |
| glyA1 | PA5415 | −10.4 ± 2.36 | Serine hydroxymethyltransferase |
| QS-related genes | |||
| lasA | PA1871 | −9.94 ± 1.45 | Protease |
| lecB | PA3361 | −34.5 ± 7.81 | Fucose-binding lectin PA-IIL |
| rhll | PA3476 | −3.59 ± 1.28 | C4-HSL synthase |
| rhlB | PA3478 | −27.4 ± 3.70 | Rhamnosyltransferase |
| rhlA | PA3479 | −38.4 ± 6.35 | Rhamnosyltransferase |
| lasB | PA3724 | −17.7 ± 2.69 | Elastase |
| Others | |||
| PA0049 | −10.3 ± 3.04 | Hypothetical | |
| rahU | PA0122 | −29.7 ± 15.5 | Aegerolysin |
| metE | PA1927 | −25.7 ± 2.09 | Methionine synthase |
| mexS | PA2491 | 12.4 ± 4.20 | Oxidoreductase |
| mexE | PA2493 | 139 ± 16.6 | Efflux membrane fusion protein |
| rahU | PA2494 | 115 ± 16.5 | Multidrug efflux transporter |
| oprN | PA2495 | 43.1 ± 8.70 | Outer membrane protein |
| PA3568 | −4.01 ± 0.46 | Probable acetyl-CoA synthetase | |
| mmsB | PA3569 | −3.42 ± 0.40 | 3-Hydroxyisobutyrate dehydrogenase |
| mmsA | PA3570 | −2.86 ± 0.42 | Methylmalonate-semialdehyde dehydrogenase |
ID, GenBank identification.
GCS, glycine cleavage system.
In sharp contrast to the overabundance of genes regulated by C4-HSL signaling identified in the microarray experiments, only a limited number of genes represent potential candidates of PA2449 regulation and are restricted to those involved in glycine metabolism (Fig. 6). On the P. aeruginosa PAO1 chromosome, the PA2449 gene is localized near a gene cluster that encodes proteins involved in glycine and serine metabolism: gcvT2 (PA2442), sdaA (PA2443), glyA2 (PA2444), gcvP2 (PA2445), and gcvH2 (PA2446). The GcvT2, GcvP2, and GcvH2 proteins constitute a glycine cleavage system used for glycine and single-carbon metabolism (51). The glyA2 gene product is predicted to be a serine hydroxymethyltransferase, and sdaA encodes a serine dehydratase that catalyzes the deamination of serine into pyruvate. All five of these proteins are necessary for glycine and serine utilization, and as shown in Fig. 6, the genes encoding these proteins were transcriptionally downregulated >10-fold in PW5126. Further analysis confirmed that the promoter region of the glycine cleavage gene gcvP2 was activated >2.5-fold in P. aeruginosa PAO1 but not in PW5126 (Fig. 7A). These data and the previous observation that PA2449 was essential for glycine utilization support a probable role for PA2449 in regulating glycine metabolism in P. aeruginosa PAO1, possibly through facilitating the transcriptional activation of the gcvT2, sdaA, glyA2, gcvP2, and gcvH2 genes.
Fig 6.
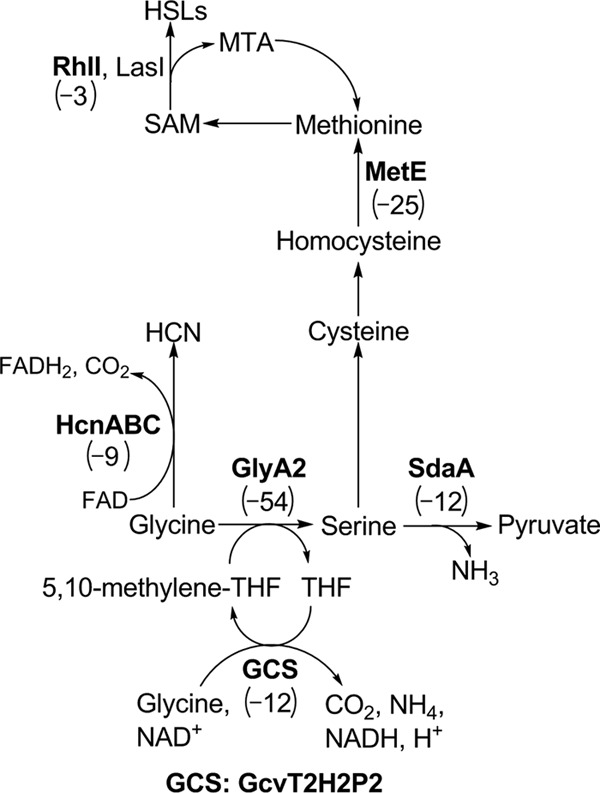
Key pathways of glycine assimilation that might be regulated by PA2449 in P. aeruginosa PAO1. Several genes whose products are predicted to be involved in glycine/serine metabolism were observed to be transcriptionally downregulated in the absence of a functional PA2449 gene. The approximate fold changes in the transcription of individual genes of interest (boldface) are given in parentheses.
Fig 7.
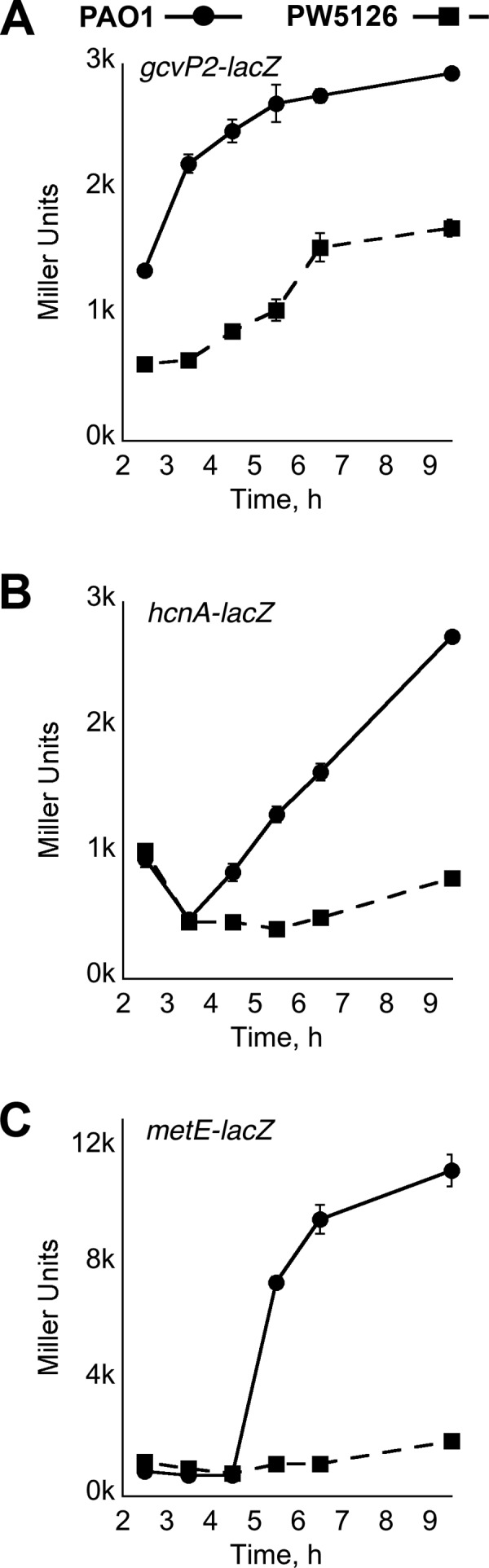
Relative expression levels of the gcvP2, hcnA, and metE genes in P. aeruginosa PAO1 and PW5126. The 5′ regulatory DNA regions consisting of ∼1,000 bp (gcvP2 [A]) or 500 bp (hcnA [B]and metE [C]) upstream of the designated gene ORF were fused with the E. coli lacZ ORF. The resulting lacZ constructs were cloned into a promoter-less pBBR1MCS-5 plasmid. P. aeruginosa strains harboring the gcvP2- (A), hcnA- (B), and metE–lacZ (C) constructs were grown in PB at 37°C and periodically assayed for LacZ activity. Individual points represent mean values of triplicate samples, and standard error bars are shown.
Other genes intimately linked to glycine metabolism in P. aeruginosa were also found to be differentially transcribed. For example, a metabolic fate of glycine in P. aeruginosa is its direct conversion into hydrogen cyanide (HCN) via the HCN synthase encoded by the hcnABC locus (52, 53). For PW5126, transcription of the hcnABC genes was down by >30-fold. A follow-up lacZ reporter assay of the hcnA promoter confirmed that its activation was PA2449 dependent (Fig. 7B). The expression of the hcnABC locus is complex, involving several different regulators such as ANR, LasR, RhlR, and GacA (53, 54). PA2449 might provide another layer of regulation, which mediates the expression of hcnABC in the presence of excess intracellular glycine in P. aeruginosa PAO1.
Glycine is also a precursor for sulfur-containing amino acids such as methionine. A gene encoding a methionine synthase, metE, was transcriptionally downregulated by 26-fold in PW5126. Examination of the metE-lacZ reporter revealed it to be activated only in P. aeruginosa PAO1 (Fig. 7C); there was a >90% decrease in expression from the metE promoter in the absence of PA2449. It is probable that PA2449 positively regulates the expression of metE to increase metabolic flux from glycine to methionine. The increase in methionine biosynthesis could lead to higher levels of S-adenoyslmethionine (SAM), thereby enhancing pyocyanin production at the PhzM catalytic step and/or the synthesis of C4-HSL by RhlI. However, the heterologous overexpression of E. coli metE or the supplementation of PB with methionine (1 to 10 mM) did not restore pyocyanin biosynthesis in PW5126 cultures. Interestingly, the expression of the metE-lacZ fusion was repressed in P. aeruginosa PAO1 PB-grown cultures that were supplemented with 1% glycine or serine but not methionine. These findings indicate that the decreased expression of metE observed from PW5126 is due to elevated intracellular concentrations of glycine/serine, which could not be sufficiently metabolized in the absence of PA2449.
In addition to a decreased capacity to consume glycine and serine, PW5126 struggled to use valine as a sole carbon source (see Fig. S3 in the supplemental material). The transcription of genes encoding a 3-hydroxyisobutyrate dehydrogenase (mmsB), methylmalonate-semialdehyde dehydrogenase (mmsA), and a propionyl-coenzyme A (propionyl-CoA) synthetase (PA3568) were downregulated >3-fold in PW5126 (see Table S4 in the supplemental material). These enzymes function in the distal pathway of valine degradation (55), and a deficiency in their expression would hinder the growth of PW5126 on valine.
Lastly, mexEF-oprN, which encodes a resistance nodulation type efflux pump, was transcriptionally upregulated by >50-fold in PW5126. This was a very unexpected result. First, MexEF-OprN overexpression has been previously observed to reduce the production of virulence factors in P. aeruginosa (56). MexEF-OprN removes intracellular PQS signaling molecules; this removal subsequently reduces the expression of rhl-related genes (57, 58). Overexpression of MexEF-OprN in PW5126 would explain the reduction in transcription of rhlI and production of C4-HSL. Second, an earlier study proposed that MexEF-OprN might cause the efflux of pyocyanin precursors (59). Coordinating the repression of the mexEF-oprN locus with the activation of genes whose products function in SAM biosynthesis, e.g., metE and gcvT2H2P2, via PA2449 could mutually enhance pyocyanin biosynthesis. Whether PA2449 functions in the repression of the mexEF-oprN locus to ensure the production of C4-HSL and pyocyanin requires further exploration.
Serine dehydratase (SdaA) is necessary for the optimal growth of P. aeruginosa PAO1 in PB.
The results of the microarray revealed several potential genes as having important roles in the production of pyocyanin and/or the survival of P. aeruginosa PAO1 in PB. In order to assess some of these genes for functionality, we requested P. aeruginosa PAO1 mutants harboring transposon insertions within the candidate genes from the PA Two Allele Library. As shown in Fig. 9, two genes were found to have a significant effect on pyocyanin production. First, disruption of the mexF gene resulted in an ∼50% increase of extracellular pyocyanin compared to that for P. aeruginosa PAO1 (Fig. 8A). This supports the role of MexEF-OprN as an antagonist in pyocyanin production. The second gene observed to impact pyocyanin yields was sdaA. There was an ∼70% reduction in pyocyanin production in the absence of the serine dehydratase gene sdaA compared to that for P. aeruginosa PAO1 (Fig. 8A). Subsequent studies showed that the sdaA mutation caused a noticeable cessation of growth in PB (Fig. 8B). This trait is similar to that of PW5126 and indicates that deamination of serine into pyruvate is an occurring metabolic process when P. aeruginosa PAO1 is grown in PB.
Fig 9.
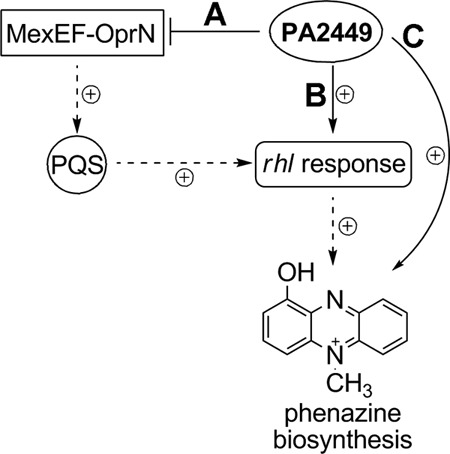
Proposed models for PA2449 in regulating the biosynthesis of C4-HSL and pyocyanin in P. aeruginosa PAO1. (A) Transcription of mexEF-oprN, encoding an efflux pump, is repressed by PA2449. This prevents the efflux of intracellular PQS, thus enhancing the expression of rhl-related genes and phenotypes, including phenazine production. (B) The PA2449 protein directly activates the transcription of the C4-HSL synthase gene, rhlI, thereby facilitating the expression of the rhl network. (C) PA2449 operates independently of the rhl network to activate the expression of phz-related genes for the biosynthesis of pyocyanin.
Fig 8.
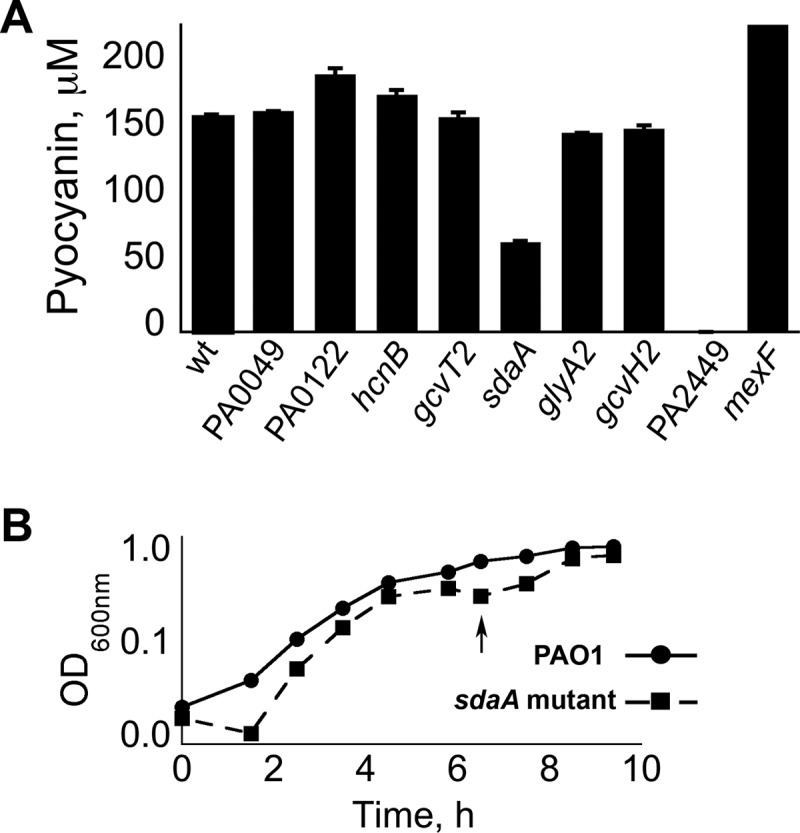
Serine dehydratase (encoded by sdaA) is necessary for the optimal growth of P. aeruginosa PAO1 in PB. (A) Pyocyanin production from P. aeruginosa mutants possessing transposon insertions within genes that are potential candidates of regulation by PA2449. Pyocyanin titers represent mean values from triplicate samples, and standard error bars are shown. wt, wild type. (B) The transposon insertion sdaA mutant has a characteristic cessation in growth (arrow) following entry into stationary phase when cultured in PB. Individual points represent mean values derived from triplicate samples, and standard error bars are shown.
DISCUSSION
PA2449 as a regulator in glycine metabolism for pseudomonads.
The PA2449 gene, which encodes a putative TyrR-like transcriptional regulator, was essential for P. aeruginosa PAO1 to assimilate glycine as a carbon source. Disruption of the PA2449 gene also reduced the growth of P. aeruginosa PAO1 on both serine and valine when either one was supplied as a carbon source. The metabolism of glycine and that of serine are interdependent on each other, so it is expected that an inability to assimilate either amino acid causes metabolic strain in the utilization of the other. In contrast, the necessity of the PA2449 gene for optimal growth on valine cannot be readily explained and remains unclear. The PA2449 gene was not required for the utilization of any amino acid, including glycine and serine, as a source of nitrogen. This suggests that a possible function of PA2449 is to aid in the metabolism of glycine as a carbon but not nitrogen source for P. aeruginosa PAO1.
Microarray analysis revealed that the disruption of the PA2449 gene affected the transcription of genes encoding proteins of a glycine cleavage system (GcvT2, GcvH2, and GcvP2), a serine hydroxymethyltransferase (GlyA2), and a serine dehydratase (SdaA). This series of five proteins enzymatically converts intracellular glycine into ammonia and pyruvate, which can then enter central metabolism (60). Deamination of serine into pyruvate was discovered to be a metabolic step in the catabolism of glycine, as disruption of the sdaA gene hindered the growth of P. aeruginosa PAO1 in a glycine-rich medium. The dissimilation of glycine into pyruvate with serine as an intermediate has been documented for other pseudomonads (60–62). Dysregulation of the glyA2 and sdaA genes would limit the metabolic flux of serine into glycine or pyruvate, respectively. glyA2 and sdaA dysregulation could account for the reduced growth of P. aeruginosa PAO1 on serine (as a carbon source) in the absence of PA2449.
Whether the PA2449 protein functions as a direct transcriptional activator for any of the genes located within the gcvT2-sdaA-glyA2-gcvP2-gcvH2 locus remains to be answered. However, the only genes related to glycine metabolism that were transcriptionally downregulated in the absence of PA2449 were gcvT2-sdaA-glyA2-gcvP2-gcvH2 and glyA1. The expression of glyA1 and glyA2 has been reported to be regulated by choline catabolism (63, 64), but we suspect that the transcriptional downregulation of glyA1 in our study was due to a deficiency in QS (65) and not a result of choline catabolism. For example, the transcription of other choline-related catabolic genes (63), including sdaB and soxBDAG, was unaffected by the disruption of the PA2449 gene. The transcription of glyA2 and gcvT2P2H2 has been shown to be influenced by QS (65) in P. aeruginosa PAO1, but neither QS nor choline catabolism has been reported to control the expression of sdaA within the gcvT2-sdaA-glyA2-gcvP2-gcvH2 locus. Lastly, the transcription of genes whose products could participate in glycine metabolism, e.g., glyA3 and gcvP1H1T1 (PA5213 to PA5215), was unaffected by the inactivation of the PA2449 gene.
PA2449 may provide another tier of transcriptional regulation of the gcvT2-sdaA-glyA2-gcvP2-gcvH2 locus. For example, PA2449 might upregulate the transcription of the gcvT2-sdaA-glyA2-gcvP2-gcvH2 locus to ensure that the resulting GcvP2H2T2-GlyA2-SdaA pathway has sufficient activity to meet the cellular demands associated with having to assimilate glycine as the only available carbon source. The PA2449 gene is conserved among pseudomonads, in which it lies in proximity to genes encoding a serine dehydratase and/or glycine cleavage system. The regulation of glycine and possibly serine catabolism by PA2449 might therefore be a universal phenomenon among Pseudomonas-related bacteria.
PA2449 is required for the biosynthesis of pyocyanin.
The production of pyocyanin in P. aeruginosa PAO1 was completely dependent on the PA2449 gene regardless of environmental conditions. Under a variety of different growth conditions and medium formulations, the PA2449 gene was essential for the production of pyocyanin. Specifically, the PA2449 gene was required for the transcriptional activation of genes, i.e., phzA1, phzA2, and phzM, necessary for the biosynthesis of pyocyanin. Without the PA2449 gene, the basal level of transcription of the phz genes was sufficient for allowing the production of PCA at trace quantities. We later found that the cause for the decreased transcription of the phz genes in the absence of PA2449 was insufficient levels of C4-HSL (discussed below), which is involved in phz gene activation.
This study characterized the relationship between the PA2449 gene and the production of pyocyanin under nutritionally poor growth conditions, i.e., glycine was the predominant source of carbon, phosphate was in limiting concentrations, and both aromatic and sulfur-containing amino acids were in scarce supply. Phosphate limitation has long been known to be a key determinant in the production of pyocyanin, but our data also implicate glycine (or a glycine-related metabolite) as a possible contributing factor for the biosynthesis of phenazines in P. aeruginosa PAO1. Glycine can be considered an unfavorable carbon source, so it is plausible that, when intracellular glycine reaches a threshold concentration under nutrient, e.g., phosphate, limitation, this might be interpreted as a starvation signal. Consequently, this could trigger the production of antimicrobials such as phenazines and HCN to ward off competitors and establish an edge for P. aeruginosa PAO1 in the environment.
We have discussed only the utilization of glycine as a carbon substrate in its relation to the production of pyocyanin, but the majority of media used for pyocyanogenic cultures do not use an exogenous source of glycine. However, limiting phosphate availability to <0.2 mM is a common formulation for enhancing pyocyanin formation (4, 29, 30). A recent transcriptome analysis of P. aeruginosa identified several genes involved in sulfate assimilation, protein biosynthesis, and purine metabolism that were downregulated under phosphate limitation (25). Because glycine (serine) is a biosynthetic precursor to thiols/purines and a product of protein turnover, a reduction in these processes could cause a gradual increase in intracellular glycine (serine) concentrations that is eventually countered by PA2449. Metabolomic analysis of P. aeruginosa PAO1 at the onset of pyocyanin production under phosphate limitation is required to validate the existence of such a scenario involving PA2449-directed glycine metabolism.
The role of PA2449 in cell signaling in P. aeruginosa PAO1.
The inactivation of the PA2449 gene significantly reduced the production of C4-HSL from P. aeruginosa PAO1. The production of pyocyanin was dependent on a C4-HSL RhlR response, as both ΔrhlR and ΔrhlR ΔlasR P. aeruginosa PAO1 mutants did not generate detectable levels of pyocyanin when grown in PB. These findings indicate that the pyocyanin-null phenotype associated with the disruption of the PA2449 gene is a result of low or insufficient levels of C4-HSL, which consequently prevent RhlR from transcriptionally activating the phz genes required for pyocyanin biosynthesis.
Although we established that the production of C4-HSL requires a functional PA2449 gene, the reasoning or mechanism(s) underlying their relationship has not yet been determined. The data presented herein show that PA2449 is necessary for the catabolism of glycine as a carbon source, but QS also has also been shown to regulate the transcription of genes involved in glycine metabolism (65). It is possible that glycine metabolism might be transcriptionally regulated only through QS in which PA2449 acts as an intermediary for facilitating the biosynthesis of C4-HSL in response to glycine availability. However, ΔrhlR, ΔlasR, and ΔrhlR ΔlasR P. aeruginosa PAO1 mutants readily grew on minimal media with glycine as the sole carbon source; a trait not shared by the transposon insertion PA2449 mutant. This suggests that glycine metabolism is dependent on PA2449 through mechanisms apart from QS in P. aeruginosa PAO1.
The necessity of the PA2449 gene, and possibly glycine metabolism, for the biosynthesis of C4-HSL is an intriguing question. One possible reason for their connection is that high intracellular levels of glycine might undesirably inhibit the production of C4-HSL. For example, glycine might be a competitive inhibitor of the RhlI synthase or the RhlR receptor. Furthermore, elevated glycine levels could interfere with the biosynthesis of homoserine and/or SAM, both of which are essential biosynthetic precursors for HSLs. In support of this mode of inhibition, we did observe that the transcription of the methionine synthase gene, metE, was repressed either by the inactivation of the PA2449 gene or through the exogenous addition of glycine/serine to P. aeruginosa PAO1 cultures. However, the addition of methionine or heterologous expression of the E. coli metE gene in the transposon insertion PA2449 mutant did not restore C4-HSL to levels sufficient for triggering the production of pyocyanin. More in-depth studies are needed to test for the effect of glycine inhibition on such physiological activities.
The reduced production of C4-HSL in the absence of the PA2449 gene could be attributed to the decreased transcription (>3-fold) of the C4-HSL synthase gene rhlI. The role of PA2449 in the transcription of rhlI was narrowed down to a few possible factors (Fig. 9). First, we observed elevated transcription (>50-fold) of the pump-encoding mexEF-oprN genes with the disruption of the PA2449 gene. Overexpression of MexEF-OprN depletes cells of intracellular PQS (56), which is needed to enhance the expression of rhl-related genes, including rhlI (46). Therefore, PA2449 might function in the repression of the mexEF-oprN locus to ensure the accumulation of intracellular PQS for activation of the rhl network and subsequent C4-HSL biosynthesis (Fig. 9A). If indeed this is the mode of regulation by PA2449 of rhlI expression, it is expected that deletion of the mexEF-oprN locus in the PA2449 mutant (PW5126) will restore intracellular PQS to levels sufficient for mediating C4-HSL biosynthesis. Closer inspection of a ΔPA2449 ΔmexEF-oprN P. aeruginosa PAO1 mutant for these physiological traits is an objective of future studies.
How PA2449 affects transcription of mexEF-oprN is unclear. An earlier study reported the direct repression of the mexEF-oprN locus from an unknown protein (66). Perhaps the PA2449 protein functions as this “unknown” repressor protein to facilitate the biosynthesis of C4-HSL under nutrient-poor conditions. In addition, the transcription of mexS (PA2493) genes was upregulated by 12-fold in the absence of PA2449. MexS participates in the expression of mexEF-oprN in response to perturbations of intracellular redox (67). An additional regulator, MvaT (PA4315), which affects the expression of several QS-related genes such as mexEF-oprN (68), was not observed to be differentially transcribed between wild-type cells and the transposon insertion PA2449 mutant. The mechanism by which PA2449 regulates the mexEF-oprN locus is currently being investigated.
In addition to influencing the transcription of the MexEF-OprN efflux pump genes, the PA2449 protein might serve as a direct transcriptional activator for the rhlI gene (Fig. 9B). The alternative sigma factor RpoN was previously found to contribute to the expression of the rhlI gene (69). Transcriptional activation of RpoN-controlled genes requires additional regulators known as enhancer-binding proteins (EBPs). The PA2449 protein is predicted to be an EBP, so it is plausible that it interacts with RpoN to mediate expression of the rhlI locus. Further in-depth studies will clarify the role of PA2449 in the production C4-HSL for P. aeruginosa PAO1.
Does the PA2449 protein function as a TyrR regulator or EBP?
This study characterizes the physiological and genetic effects of a transposon deletion PA2449 mutant of P. aeruginosa PAO1. The PA2449 gene is predicted to encode either a TyrR or EBP type transcriptional regulator. This ambiguity is due to the fact that the PA2449 protein possess homology to both the TyrR protein of E. coli and the EBP PhhR of P. aeruginosa. However, an alignment between these three proteins shows that the primary sequence of the PA2449 protein is 44% identical to that of PhhR and 41% identical to that of E. coli TyrR. Additionally, there is a stretch of ∼8 amino acid residues unique to only the PA2449 protein and PhhR. These amino acid residues are located within the predicated RpoN interaction domain, which would explain their absence from the non-EBP TyrR regulator of E. coli. Several of the genes that are potentially regulated by the PA2449 protein, including gcvH2, phzB1, and rhlI, possess putative RpoN promoters (http://www.sigma54.ca/promoterdata/Web/data.aspx). We were unable to culture a ΔrpoN P. aeruginosa PAO1 strain in peptone broth, stressing the importance RpoN-mediated transcription under nutrient-poor conditions. Additionally, RpoN has been reported to be essential for the utilization of glycine/serine as carbon and nitrogen sources in pseudomonads (70, 71). These data support a probable role for the PA2449 protein as an EBP that interacts with RpoN to mediate transcription under conditions in which glycine is the predominant carbon source.
To biochemically characterize and verify that the PA2449 protein behaves as an EBP, it is imperative to determine the intracellular signal(s) sensed by this protein. Our data clearly show that PA2449 is involved in glycine metabolism, so an obvious candidate for triggering the PA2449 protein into an active state is glycine or possibly its metabolic counterpart serine. Furthermore, excess intracellular concentrations of glycine are expected to have adverse effects on the availability of single-carbon carrier molecules of the tetrahydrofolate (THF) family, e.g., 5,10-methenyl-THF (51). The PA2449 protein may interact with a specific THF derivative, thus activating expression of GcvP2H2T2-GlyA2-SdaA and subsequently restoring balance to the THF pool. PA2449 was involved in C4-HSL biosynthesis by possibly augmenting the PQS response. Does the PA2449 protein respond to PQS? We are currently examining the biochemical function of the PA2449 protein and attempting to define its in vivo targets in P. aeruginosa PAO1 when grown under glycine-rich, pyocyanin-producing conditions.
Supplementary Material
ACKNOWLEDGMENTS
We thank Linda Thomashow (USDA) for supplying us with an authentic phenazine-1-carboxylic acid and the pUCP-A2G2. The ΔrhlR, ΔlasR and ΔrhlR ΔlasR P. aeruginosa PAO1 strains were kindly provided by E. Peter Greenberg (University of Washington). We thank Karen Gentile and Frank Middleton from Microarray Core Facility of SUNY Upstate for assistance. We thank Ammar Saleem (University of Ottawa) for assistance with HPLC.
We acknowledge grant NIH P30 DK089507 for funding the P. aeruginosa PAO1 transposon mutants used for our study. This study was made possible by NSF DMR 090785 and USDA CSREES awards to C. T. Nomura.
Footnotes
Published ahead of print 1 March 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.02205-12.
REFERENCES
- 1. Laursen JB, Nielsen J. 2004. Phenazine natural products: biosynthesis, synthetic analogues, and biological activity. Chem. Rev. 104:1663–1686 [DOI] [PubMed] [Google Scholar]
- 2. Price-Whelan A, Dietrich LE, Newman DK. 2006. Rethinking ‘secondary’ metabolism: physiological roles for phenazine antibiotics. Nat. Chem. Biol. 2:71–78 [DOI] [PubMed] [Google Scholar]
- 3. Wang Y, Wilks JC, Danhorn T, Ramos I, Croal L, Newman DK. 2011. Phenazine-1-carboxylic acid promotes bacterial biofilm development via ferrous iron acquisition. J. Bacteriol. 193:3606–3617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cox CD. 1986. Role of pyocyanin in the acquisition of iron from transferrin. Infect. Immun. 52:263–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Price-Whelan A, Dietrich LE, Newman DK. 2007. Pyocyanin alters redox homeostasis and carbon flux through central metabolic pathways in Pseudomonas aeruginosa PA14. J. Bacteriol. 189:6372–6381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wang Y, Kern SE, Newman DK. 2010. Endogenous phenazine antibiotics promote anaerobic survival of Pseudomonas aeruginosa via extracellular electron transfer. J. Bacteriol. 192:365–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dietrich LE, Price-Whelan A, Petersen A, Whiteley M, Newman DK. 2006. The phenazine pyocyanin is a terminal signalling factor in the quorum sensing network of Pseudomonas aeruginosa. Mol. Microbiol. 61:1308–1321 [DOI] [PubMed] [Google Scholar]
- 8. D'Aes J, Hua GK, De Maeyer K, Pannecoucque J, Forrez I, Ongena M, Dietrich LE, Thomashow LS, Mavrodi DV, Hofte M. 2011. Biological control of Rhizoctonia root rot on bean by phenazine- and cyclic lipopeptide-producing Pseudomonas CMR12a. Phytopathology 101:996–1004 [DOI] [PubMed] [Google Scholar]
- 9.. Mavrodi DV, Parejko JA, Mavrodi OV, Kwak YS, Weller DM, Blankenfeldt W, Thomashow LS. Recent insights into the diversity, frequency and ecological roles of phenazines in fluorescent Pseudomonas spp. Environ. Microbiol., in press [DOI] [PubMed] [Google Scholar]
- 10. Weller DM, Raaijmakers JM, Gardener BB, Thomashow LS. 2002. Microbial populations responsible for specific soil suppressiveness to plant pathogens. Annu. Rev. Phytopathol. 40:309–348 [DOI] [PubMed] [Google Scholar]
- 11. Denning GM, Iyer SS, Reszka KJ, O'Malley Y, Rasmussen GT, Britigan BE. 2003. Phenazine-1-carboxylic acid, a secondary metabolite of Pseudomonas aeruginosa, alters expression of immunomodulatory proteins by human airway epithelial cells. Am. J. Physiol. Lung Cell. Mol. Physiol. 285:L584–L592 [DOI] [PubMed] [Google Scholar]
- 12. Lau GW, Hassett DJ, Ran H, Kong F. 2004. The role of pyocyanin in Pseudomonas aeruginosa infection. Trends Mol. Med. 10:599–606 [DOI] [PubMed] [Google Scholar]
- 13. Lau GW, Ran H, Kong F, Hassett DJ, Mavrodi D. 2004. Pseudomonas aeruginosa pyocyanin is critical for lung infection in mice. Infect. Immun. 72:4275–4278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wilson R, Pitt T, Taylor G, Watson D, MacDermot J, Sykes D, Roberts D, Cole P. 1987. Pyocyanin and 1-hydroxyphenazine produced by Pseudomonas aeruginosa inhibit the beating of human respiratory cilia in vitro. J. Clin. Invest. 79:221–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wilson R, Sykes DA, Watson D, Rutman A, Taylor GW, Cole PJ. 1988. Measurement of Pseudomonas aeruginosa phenazine pigments in sputum and assessment of their contribution to sputum sol toxicity for respiratory epithelium. Infect. Immun. 56:2515–2517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mavrodi DV, Bonsall RF, Delaney SM, Soule MJ, Phillips G, Thomashow LS. 2001. Functional analysis of genes for biosynthesis of pyocyanin and phenazine-1-carboxamide from Pseudomonas aeruginosa PAO1. J. Bacteriol. 183:6454–6465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mentel M, Ahuja EG, Mavrodi DV, Breinbauer R, Thomashow LS, Blankenfeldt W. 2009. Of two make one: the biosynthesis of phenazines. Chembiochem 10:2295–2304 [DOI] [PubMed] [Google Scholar]
- 18. Mavrodi DV, Ksenzenko VN, Bonsall RF, Cook RJ, Boronin AM, Thomashow LS. 1998. A seven-gene locus for synthesis of phenazine-1-carboxylic acid by Pseudomonas fluorescens 2-79. J. Bacteriol. 180:2541–2548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Whiteley M, Lee KM, Greenberg EP. 1999. Identification of genes controlled by quorum sensing in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 96:13904–13909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chugani SA, Whiteley M, Lee KM, D'Argenio D, Manoil C, Greenberg EP. 2001. QscR, a modulator of quorum-sensing signal synthesis and virulence in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 98:2752–2757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schuster M, Greenberg EP. 2006. A network of networks: quorum-sensing gene regulation in Pseudomonas aeruginosa. Int. J. Med. Microbiol. 296:73–81 [DOI] [PubMed] [Google Scholar]
- 22. Pearson JP, Passador L, Iglewski BH, Greenberg EP. 1995. A second N-acylhomoserine lactone signal produced by Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 92:1490–1494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pearson JP, Gray KM, Passador L, Tucker KD, Eberhard A, Iglewski BH, Greenberg EP. 1994. Structure of the autoinducer required for expression of Pseudomonas aeruginosa virulence genes. Proc. Natl. Acad. Sci. U. S. A. 91:197–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pesci EC, Milbank JB, Pearson JP, McKnight S, Kende AS, Greenberg EP, Iglewski BH. 1999. Quinolone signaling in the cell-to-cell communication system of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 96:11229–11234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bains M, Fernandez L, Hancock RE. 2012. Phosphate starvation promotes swarming motility and cytotoxicity of Pseudomonas aeruginosa. Appl. Environ. Microbiol. 78:6762–6768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Long J, Zaborina O, Holbrook C, Zaborin A, Alverdy J. 2008. Depletion of intestinal phosphate after operative injury activates the virulence of Pseudomonas aeruginosa causing lethal gut-derived sepsis. Surgery 144:189–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Frank LH, Demoss RD. 1959. On the biosynthesis of pyocyanine. J. Bacteriol. 77:776–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Oglesby AG, Farrow JM, III, Lee JH, Tomaras AP, Greenberg EP, Pesci EC, Vasil ML. 2008. The influence of iron on Pseudomonas aeruginosa physiology: a regulatory link between iron and quorum sensing. J. Biol. Chem. 283:15558–15567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Essar DW, Eberly L, Hadero A, Crawford IP. 1990. Identification and characterization of genes for a second anthranilate synthase in Pseudomonas aeruginosa: interchangeability of the two anthranilate synthases and evolutionary implications. J. Bacteriol. 172:884–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Macdonald JC. 1963. Biosynthesis of pyocyanine. Can. J. Microbiol. 9:809–819 [Google Scholar]
- 31. Heurlier K, Denervaud V, Haenni M, Guy L, Krishnapillai V, Haas D. 2005. Quorum-sensing-negative (lasR) mutants of Pseudomonas aeruginosa avoid cell lysis and death. J. Bacteriol. 187:4875–4883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Miller JH. 1972. Experiments in molecular genetics, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 33. Jacobs MA, Alwood A, Thaipisuttikul I, Spencer D, Haugen E, Ernst S, Will O, Kaul R, Raymond C, Levy R, Chun-Rong L, Guenthner D, Bovee D, Olson MV, Manoil C. 2003. Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 100:14339–14344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Choi KH, Kumar A, Schweizer HP. 2006. A 10-min method for preparation of highly electrocompetent Pseudomonas aeruginosa cells: application for DNA fragment transfer between chromosomes and plasmid transformation. J. Microbiol. Methods 64:391–397 [DOI] [PubMed] [Google Scholar]
- 35. Szewczyk E, Nayak T, Oakley CE, Edgerton H, Xiong Y, Taheri-Talesh N, Osmani SA, Oakley BR. 2006. Fusion PCR and gene targeting in Aspergillus nidulans. Nat. Protoc. 1:3111–3120 [DOI] [PubMed] [Google Scholar]
- 36. Zhang X, Bremer H. 1995. Control of the Escherichia coli rrnB P1 promoter strength by ppGpp. J. Biol. Chem. 270:11181–11189 [DOI] [PubMed] [Google Scholar]
- 37. Trutko SM, Garagulya AD, Kiprianova EA, Akimenko VK. 1989. The physiological role of phenazine pigments synthesized by the bacteria Pseudomonas aureofaciens. Biochemistry (Mosc) 54:1092–1098 [Google Scholar]
- 38. Henderson JW, Ricker RD, Bidlingmeyer BA, Woodward C. 2000. Rapid, accurate, sensitive, and reproducible HPLC analysis of amino acids. AAA technical note P5 publication no. 5980-1193. Agilent Technologies, Santa Clara, CA [Google Scholar]
- 39. MacEachran DP, Ye S, Bomberger JM, Hogan DA, Swiatecka-Urban A, Stanton BA, O'Toole GA. 2007. The Pseudomonas aeruginosa secreted protein PA2934 decreases apical membrane expression of the cystic fibrosis transmembrane conductance regulator. Infect. Immun. 75:3902–3912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Morett E, Segovia L. 1993. The sigma 54 bacterial enhancer-binding protein family: mechanism of action and phylogenetic relationship of their functional domains. J. Bacteriol. 175:6067–6074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Song J, Jensen RA. 1996. PhhR, a divergently transcribed activator of the phenylalanine hydroxylase gene cluster of Pseudomonas aeruginosa. Mol. Microbiol. 22:497–507 [DOI] [PubMed] [Google Scholar]
- 42. Palmer GC, Palmer KL, Jorth PA, Whiteley M. 2010. Characterization of the Pseudomonas aeruginosa transcriptional response to phenylalanine and tyrosine. J. Bacteriol. 192:2722–2728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Palmer KL, Aye LM, Whiteley M. 2007. Nutritional cues control Pseudomonas aeruginosa multicellular behavior in cystic fibrosis sputum. J. Bacteriol. 189:8079–8087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. King EO, Ward MK, Raney DE. 1954. Two simple media for the demonstration of pyocyanin and fluorescin. J. Lab. Clin. Med. 44:301–307 [PubMed] [Google Scholar]
- 45. Cataldi TR, Bianco G, Abate S. 2009. Accurate mass analysis of N-acyl-homoserine-lactones and cognate lactone-opened compounds in bacterial isolates of Pseudomonas aeruginosa PAO1 by LC-ESI-LTQ-FTICR-MS. J. Mass Spectrom. 44:182–192 [DOI] [PubMed] [Google Scholar]
- 46. McKnight SL, Iglewski BH, Pesci EC. 2000. The Pseudomonas quinolone signal regulates rhl quorum sensing in Pseudomonas aeruginosa. J. Bacteriol. 182:2702–2708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mashburn LM, Jett AM, Akins DR, Whiteley M. 2005. Staphylococcus aureus serves as an iron source for Pseudomonas aeruginosa during in vivo coculture. J. Bacteriol. 187:554–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Deziel E, Lepine F, Milot S, He J, Mindrinos MN, Tompkins RG, Rahme LG. 2004. Analysis of Pseudomonas aeruginosa 4-hydroxy-2-alkylquinolines (HAQs) reveals a role for 4-hydroxy-2-heptylquinoline in cell-to-cell communication. Proc. Natl. Acad. Sci. U. S. A. 101:1339–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Farrow JM, Pesci EC. 2007. Two distinct pathways supply anthranilate as a precursor of the Pseudomonas quinolone signal. J. Bacteriol. 189:3425–3433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Gray VL, O'Reilly M, Muller CT, Watkins ID, Lloyd D. 2006. Low tyrosine content of growth media yields aflagellate Salmonella enterica serovar Typhimurium. Microbiology 152:23–28 [DOI] [PubMed] [Google Scholar]
- 51. Kikuchi G, Motokawa Y, Yoshida T, Hiraga K. 2008. Glycine cleavage system: reaction mechanism, physiological significance, and hyperglycinemia. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 84:246–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Castric PA. 1977. Glycine metabolism by Pseudomonas aeruginosa: hydrogen cyanide biosynthesis. J. Bacteriol. 130:826–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Pessi G, Haas D. 2000. Transcriptional control of the hydrogen cyanide biosynthetic genes hcnABC by the anaerobic regulator ANR and the quorum-sensing regulators LasR and RhlR in Pseudomonas aeruginosa. J. Bacteriol. 182:6940–6949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Pessi G, Haas D. 2001. Dual control of hydrogen cyanide biosynthesis by the global activator GacA in Pseudomonas aeruginosa PAO1. FEMS Microbiol. Lett. 200:73–78 [DOI] [PubMed] [Google Scholar]
- 55. Steele MI, Lorenz D, Hatter K, Park A, Sokatch JR. 1992. Characterization of the mmsAB operon of Pseudomonas aeruginosa PAO encoding methylmalonate-semialdehyde dehydrogenase and 3-hydroxyisobutyrate dehydrogenase. J. Biol. Chem. 267:13585–13592 [PubMed] [Google Scholar]
- 56. Köhler T, van Delden C, Curty LK, Hamzehpour MM, Pechere JC. 2001. Overexpression of the MexEF-OprN multidrug efflux system affects cell-to-cell signaling in Pseudomonas aeruginosa. J. Bacteriol. 183:5213–5222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Olivares J, Alvarez-Ortega C, Linares JF, Rojo F, Köhler T, Martinez JL. 2012. Overproduction of the multidrug efflux pump MexEF-OprN does not impair Pseudomonas aeruginosa fitness in competition tests, but produces specific changes in bacterial regulatory networks. Environ. Microbiol. 14:1968–1981 [DOI] [PubMed] [Google Scholar]
- 58. Lamarche MG, Deziel E. 2011. MexEF-OprN efflux pump exports the Pseudomonas quinolone signal (PQS) precursor HHQ (4-hydroxy-2-heptylquinoline). PLoS One 6:e24310 doi:10.1371/journal.pone.0024310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Köhler T, Michea-Hamzehpour M, Henze U, Gotoh N, Curty LK, Pechere JC. 1997. Characterization of MexE-MexF-OprN, a positively regulated multidrug efflux system of Pseudomonas aeruginosa. Mol. Microbiol. 23:345–354 [DOI] [PubMed] [Google Scholar]
- 60. Wong HC, Lessie TG. 1979. Hydroxy amino acid metabolism in Pseudomonas cepacia: role of L-serine deaminase in dissimilation of serine, glycine, and threonine. J. Bacteriol. 140:240–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Jacob GS, Garbow JR, Schaefer J, Kishore GM. 1987. Solid-state NMR studies of regulation of N-(phosphonomethyl)glycine and glycine metabolism in Pseudomonas sp. strain PG2982. J. Biol. Chem. 262:1552–1557 [PubMed] [Google Scholar]
- 62. Kishore GM, Jacob GS. 1987. Degradation of glyphosate by Pseudomonas sp. PG2982 via a sarcosine intermediate. J. Biol. Chem. 262:12164–12168 [PubMed] [Google Scholar]
- 63. Diab F, Bernard T, Bazire A, Haras D, Blanco C, Jebbar M. 2006. Succinate-mediated catabolite repression control on the production of glycine betaine catabolic enzymes in Pseudomonas aeruginosa PAO1 under low and elevated salinities. Microbiology 152:1395–1406 [DOI] [PubMed] [Google Scholar]
- 64. Wargo MJ, Szwergold BS, Hogan DA. 2008. Identification of two gene clusters and a transcriptional regulator required for Pseudomonas aeruginosa glycine betaine catabolism. J. Bacteriol. 190:2690–2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Schuster M, Lostroh CP, Ogi T, Greenberg EP. 2003. Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: a transcriptome analysis. J. Bacteriol. 185:2066–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Maseda H, Uwate M, Nakae T. 2010. Transcriptional regulation of the mexEF-oprN multidrug efflux pump operon by MexT and an unidentified repressor in nfxC-type mutant of Pseudomonas aeruginosa. FEMS Microbiol. Lett. 311:36–43 [DOI] [PubMed] [Google Scholar]
- 67. Fargier E, Mac Aogain M, Mooij MJ, Woods DF, Morrissey JP, Dobson AD, Adams C, O'Gara F. 2012. MexT functions as a redox-responsive regulator modulating disulfide stress resistance in Pseudomonas aeruginosa. J. Bacteriol. 194:3502–3511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Westfall LW, Carty NL, Layland N, Kuan P, Colmer-Hamood JA, Hamood AN. 2006. mvaT mutation modifies the expression of the Pseudomonas aeruginosa multidrug efflux operon mexEF-oprN. FEMS Microbiol. Lett. 255:247–254 [DOI] [PubMed] [Google Scholar]
- 69. Thompson LS, Webb JS, Rice SA, Kjelleberg S. 2003. The alternative sigma factor RpoN regulates the quorum sensing gene rhlI in Pseudomonas aeruginosa. FEMS Microbiol. Lett. 220:187–195 [DOI] [PubMed] [Google Scholar]
- 70. Köhler T, Harayama S, Ramos JL, Timmis KN. 1989. Involvement of Pseudomonas putida RpoN sigma factor in regulation of various metabolic functions. J. Bacteriol. 171:4326–4333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Li W, Lu CD. 2007. Regulation of carbon and nitrogen utilization by CbrAB and NtrBC two-component systems in Pseudomonas aeruginosa. J. Bacteriol. 189:5413–5420 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.