Table 3. The 37 Molecules Tested for Binding to L99A/M102H†a.
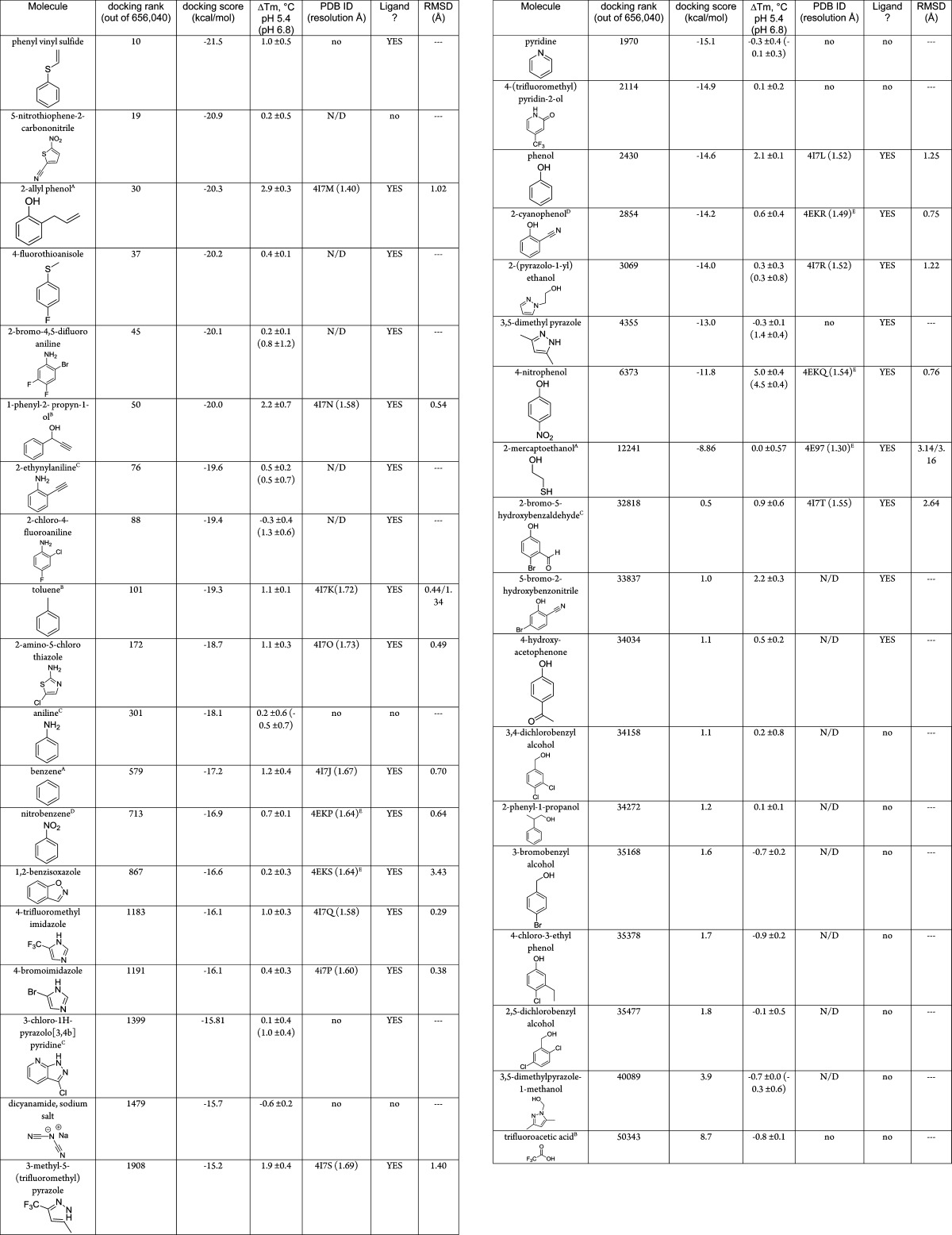
Molecules were identified as ligands if they showed a thermal unfolding upshift ΔTm of ≥0.4 °C in the presence of 1 mM ligand, unless otherwise noted, at pH 5.4, pH 6.8, or pH 3.05, or a co-crystal structure was successfully obtained (at pH 4.5). Those molecules which a crystal complex structure could not be obtained by soaking are indicated by “no” in the PDB ID column, those ligands that were not screened this way are reported as “N/D”. A more thorough treatment is included as Supporting Information Tables 2 and 3. The top 10000 molecules in the docking ranked list are included as Supporting Information Table 4. A = tested at 2 mM. B = tested at 5 mM. C = tested at 0.5 mM. D = measured at pH = 3.05. E = previously reported.37
