Table 1.
Summary of the detected hotspots
| Hotspot index | Sizea | Locib | GO categoryc | Hitsd | t-test (all)e | t-test (hits)f |
|---|---|---|---|---|---|---|
| NO. 1 | 32 | Chr XII:1056103 | Telomere maintenance via recombination
|
5 | 20 | 5 |
| NO. 2 | 27 | Chr IV:1525327 | Telomere maintenance via recombination
|
4 | 5 | 3 |
| NO. 3 | 26 | Chr XII:662627 | Sterol metabolic process
|
7 | 25 | 7 |
| NO. 4 | 24 | Chr I:52859 | Fatty acid metabolic process
|
10 | 12 | 6 |
| NO. 5 | 24 | Chr XV:202370 | Response to abiotic stimulus
|
10 | 11 | 6 |
| NO. 6 | 23 | Chr III:201166 | Response to pheromone
|
7 | 16 | 6 |
| NO. 7 | 21 | Chr VII:402833 | Protein folding
|
8 | 4 | 3 |
| NO. 8 | 19 | Chr I:7298 | Fatty acid beta-oxidation
|
5 | 13 | 4 |
| NO. 9 | 18 | Chr IV:33214 |
Response to toxin
|
5 | 0 | 0 |
| NO. 10 | 16 | Chr II:562415 | Cytokinesis
|
8 | 15 | 8 |
| NO. 11 | 16 | Chr X:698149 | — | — | 3 | — |
| NO. 12 | 16 | Chr XV:132423 | — | — | 13 | — |
| NO. 13 | 15 | Chr XIII:843356 |
Organic acid transport
|
5 | 0 | 0 |
| NO. 14 | 15 | Chr V:395442 | — | — | 3 | — |
| NO. 15 | 15 | Chr XVI:486637 | Sexual reproduction
|
6 | 8 | 4 |
aNumber of genes associated with the hotspot. bThe chromosome position of hotspot. cThe most significant GO category enriched in the associated gene set. The enrichment test was performed using DAVID (Huang da and Lempicki, 2008). The gene function is defined by GO Fat category. DAVID outputs the Benjamini–Hochberg adjusted P-value. Adjusted P-values are indicated by  , where
, where 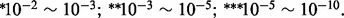 dNumber of associated genes that are functional in the enriched GO category. eNumber of associated genes that can also be identified using t-test. fNumber of associated genes that are functional in the enriched GO category and can also be identified using t-test. Two novel hotspots (NO. 9 and NO. 13) which cannot be detected by standard linear regression are in bold.
dNumber of associated genes that are functional in the enriched GO category. eNumber of associated genes that can also be identified using t-test. fNumber of associated genes that are functional in the enriched GO category and can also be identified using t-test. Two novel hotspots (NO. 9 and NO. 13) which cannot be detected by standard linear regression are in bold.
