Fig. 1.
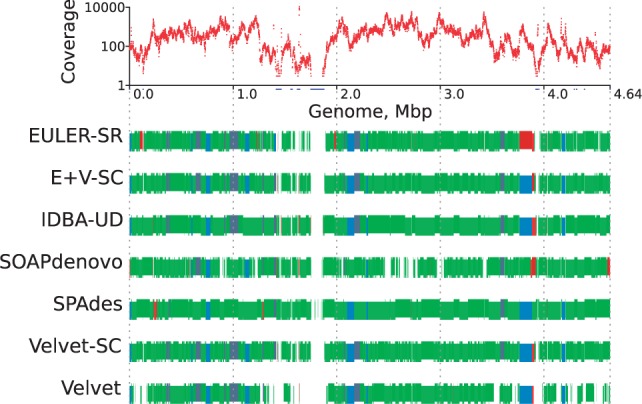
Alignment of single-cell E.coli assemblies to the reference genome. On all tracks, the x-axis is genome position. Top track: Read coverage on a logarithmic scale. The red curve shows coverage binned in 1000 bp windows. Blue positions on the x-axis have zero coverage, even if their bin has some coverage. Coverage is highly non-uniform, ranging from 0 to near 10 000. All other tracks: Comparison of positions of aligned contigs. Contigs that align correctly are coloured blue if the boundaries agree (within 2000 bp on each side) in at least half of the assemblies, and green otherwise. Contigs with misassemblies are broken into blocks and coloured orange if the boundaries agree in at least half of the assemblies, and red otherwise. Contigs are staggered vertically and are shown in different shades of their colour to distinguish the separate contigs, including small ones
