Table 1.
Comparison of assemblies of a single-cell sample of E.coli (for contigs 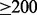 bp)
bp)
| Assembler | No. of contigs | NGA50 (bp) | Largest (bp) | Total (bp) | Genome fraction (%) | No. of misassemblies | No. of complete genes |
|---|---|---|---|---|---|---|---|
| EULER-SR | 610 | 26 580 | 140 518 | 4 306 898 | 86.54 | 19 | 3442 |
| E+V-SC | 396 | 32 051 | 132 865 | 4 555 721 | 93.58 | 2 | 3816 |
| IDBA-UD | 283 | 90 607 | 224 018 | 4 734 432 | 95.90 | 9 | 4030 |
| SOAPdenovo | 817 | 16 606 | 87 533 | 4 183 037 | 81.36 | 6 | 3060 |
| SPAdes | 532 | 99 913 | 211 020 | 4 975 641 | 96.99 | 11 | 4071 |
| Velvet | 310 | 22 648 | 132 865 | 3 517 182 | 75.53 | 2 | 3121 |
| Velvet-SC | 617 | 19 791 | 121 367 | 4 556 809 | 93.31 | 2 | 3662 |
The best value for each column is indicated in bold.
