Fig. 1.
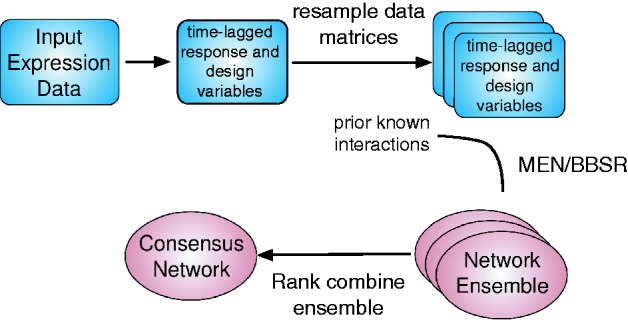
Method flow chart. Our method takes as input an expression dataset. To build a mechanistic model of gene expression, we create time-lagged response and design variables, such that the expression of the TF is time-lagged with respect to the expression of the target. We then resample the response and designing matrices, running model selection (using either MEN or BBSR) for each resample. This generates an ensemble of networks, which we rank combine into one final network
