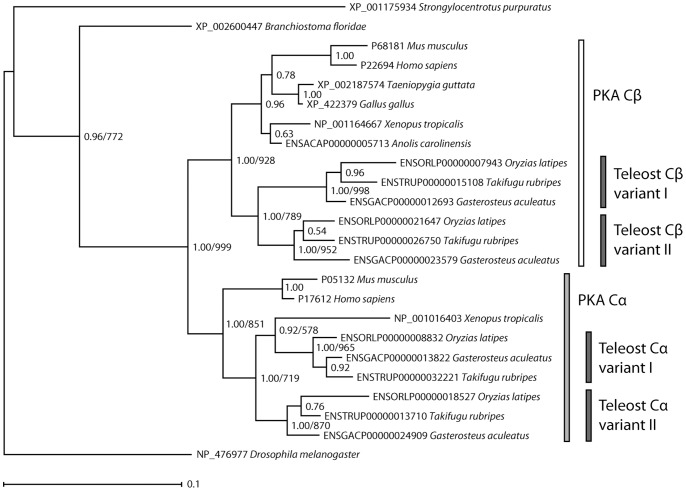
Figure 2. Phylogenetic relationships among the PKA catalytic subunit homologs in chordates.
The Cα and Cβ paralogs are a result of a gene duplication in a common ancestor of vertebrates. Subsequent duplications of Cα and Cβ in a teleost fish ancestor have resulted in four PKA catalytic subunits in these organisms. The Bayesian inference tree is based on the nucleotide sequences (codon positions 1 and 2 only, GTR+Γ+I model) of exons 2 to 10 which corresponds to a multiple sequence alignment with no gaps. The phylogram is shown with estimated branch lengths proportional to the number of substitutions at each site, as indicated by the scale bar. The arthropod fruit fly (D. melanogaster) and the echinoderm sea urchin (S. purpuratus) have been set as outgroups. Bayesian posterior probabilities are shown for each node. The topology of a maximum likelihood (ML) tree generated with the same data set and model was identical to the Bayesian inference tree. ML bootstrap values are shown for selected nodes (1000 replications). The sequences of human and mouse PKA Cα and Cβ and the homologs from amphioxus (B. floridae), zebra finch (T. guttata), chicken (G. gallus), the frog X. tropicalis, the lizard A. carolinensis, medaka (O. latipes), the pufferfish T. rubripes, and stickleback (G. aculeatus) are described in Materials and Methods S1. The X. tropicalis Cα and A. carolinensis Cβ are incorrectly placed (See discussion and Fig. 3).