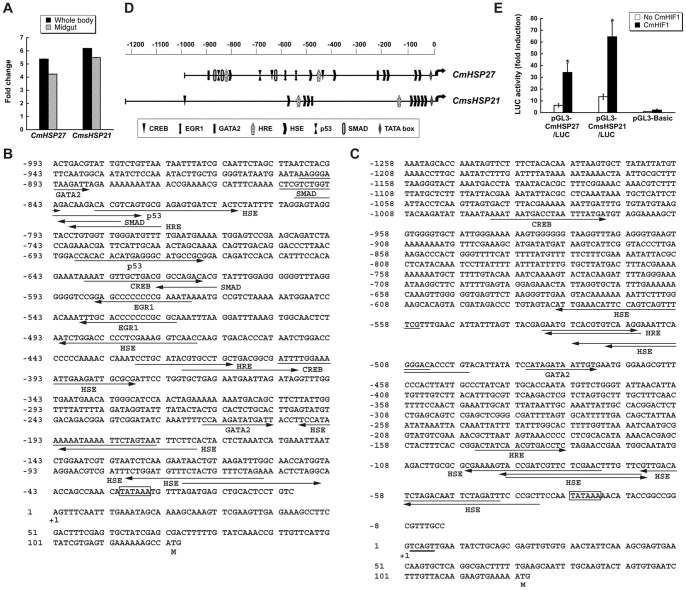
Figure 5. CmHIF1 enhances the promoter activity of selected CmHSPs.
(A) Cowpea bruchids induce expression of CmHSP27 and CmsHSP21 in response to hypoxia/hypercapnia. After 24 hr exposure, total RNA was extracted from whole body and midgut, respectively. qRT-PCR was performed to profile gene expression of these selected heat shock proteins. (B, C) Architecture of genomic DNA upstream of CmHSP27 (B) and CmsHSP21 (C) coding regions. Transcription initiation site is marked as +1, and the upstream sequences are denoted with negative numbers. Potential cis-elements are illustrated by arrows under DNA sequence in putative CmHSP27 and CmsHSP21 promoters. Putative TATA boxes are boxed, and the pentamer sequence is underlined. (D) Schematic comparison of the promoter regions of CmHSP27 and CmsHSP21 genes. Both promoters have binding sites for HIF1, CREB, GATA2, HSF family and TATA box. (E) CmHIF1 enhances the promoter activity of CmHSPs in Drosophila S2 cells. S2 cells were cotransfected with 8 μg of LUC reporter plasmids and 0.5 μg of each expression plasmid (pAc5-CmHIF1α and pAc5-CmHIF1β; black bars) or equivalent empty expression vector (white bars), respectively. The latter was used to ensure comparable total DNA amounts in transfected S2 cells. The difference between LUC activity of reporter plasmids, pGL3-CmHSP27/LUC and pGL3-CmsHSP21/LUC showed statistical significance (t test, p<0.05) in the presence versus absence of CmHIF1. The reporter plasmid pGL3-Basic was included as a control for LUC activity. Renilla luciferase vector (pRL-SV40) was used as an internal control for transfection efficiency. Luciferase activity is expressed as fold induction relative to that of the pGL3-Basic vector. Error bars represent the means ± S.E. of four independent cotransfections.