Abstract
The protective efficacy of recombinant vaccines expressing serotype 8 bluetongue virus (BTV-8) capsid proteins was tested in a mouse model. The recombinant vaccines comprised plasmid DNA or Modified Vaccinia Ankara viruses encoding BTV VP2, VP5 or VP7 proteins. These constructs were administered alone or in combination using either a homologous prime boost vaccination regime (rMVA/rMVA) or a heterologous vaccination regime (DNA/rMVA). The DNA/rMVA or rMVA/rMVA prime-boost were administered at a three week interval and all of the animals that received VP2 generated neutralising antibodies. The vaccinated and non-vaccinated-control mice were subsequently challenged with a lethal dose of BTV-8. Mice vaccinated with VP7 alone were not protected. However, mice vaccinated with DNA/rMVA or rMVA/rMVA expressing VP2, VP5 and VP7 or VP2 alone were all protected.
Introduction
Bluetongue virus (BTV) is an arthropod borne, non-enveloped virus of the genus Orbivirus, within the family Reoviridae. It is transmitted by biting midges of the genus Culicoides and infects ruminants causing severe haemorrhagic ‘bluetongue’ disease (BT) particularly in sheep and some species of deer [1], [2]. The BTV genome is composed of ten linear segments of dsRNA encoding seven structural and four distinct non-structural virus proteins (VP1–VP7 and NS1–NS4 respectively) [3], [4]. The genome segments are packaged within a three-layered icosahedral protein capsid [5], [6], [7], [8], [9]. The BTV outer-capsid layer is composed of VP2 and VP5 proteins, encoded by genome segments 2 and 6 (Seg-2 and Seg-6) respectively. The outer-core layer is formed by VP7 protein, encoded by Seg-7, while the inner-most sub-core shell is formed of VP3 protein, encoded by Seg-3 [3], [7].
VP2 is the most variable of the BTV proteins and is a major protective antigen. The specificity of its interactions with neutralising antibodies determines the identity of the 26 known BTV serotypes [6], [10], [11], [12]. Consequently, differences in the amino acid sequence of VP2 show a close correlation with virus serotype [10]. However, there are also differences within each serotype that reflect the geographic origin (topotype) of the virus isolate [11], [12].
Although the smaller BTV outer-capsid protein VP5 is also highly variable, its sequence only shows a partial correlation with virus serotype and VP5 by itself does not appear to raise neutralising antibodies [6], [13]. However, although studies of BTV neutralisation-escape mutants mostly showed changes in VP2, such changes were occasionally also observed in VP5 [14]. Studies of reassortant progeny viruses, derived from parental strains belonging to two different BTV serotypes, suggest that interactions between VP2 and VP5 can affect the serological properties of the virus, possibly by VP5 influencing the conformation of VP2 [15], [16]. BTV outer-core protein VP7 does not appear to be exposed on the surface of intact bluetongue virus-particles [17], although it can mediate both cell attachment and penetration by BTV core-particles during the initial stages of infection of insect cells or adult vector insects [18]. Antibodies to VP7 can also bind to and neutralise core particles, but do not reduce the infectivity of the intact virus [17].
Since 1998, BT outbreaks have spread across the entire Mediterranean region, caused by BTV serotypes 1, 2, 4, 6, 8, 9, 11, 16 and 25, in some cases involving more than one strain or ‘topotype’ of the same serotype [19], [20]. The first BT outbreak ever recorded in northern Europe, started during 2006, caused by a ‘western’ strain of BTV-8 from sub-Saharan Africa [11]. The outbreak was first reported in the Maastricht region of the Netherlands, although it may have started earlier the same year in Belgium [21]. From this initial introduction, BTV-8 spread across most of Europe, killing many thousands of animals and causing massive economic losses (European Commission. Restriction zones of bluetongue in Europe as of December 19, 2007, cited 2007 December 27, Available from http://ec.europa.eu/food/animal/diseases/controlmeasures/bluetongue_en.htm). The virus arrived in the UK during August 2007, transmitted by wind-borne infected midges from the outbreak regions on the European mainland [22]. Although initial control measures, relied primarily on restriction of animal movements from the BTV-8 infected areas, the use of an inactivated vaccine in the UK during early 2008 prevented the re-emergence of the disease. Subsequent vaccination campaigns in other northern European countries (France, Belgium, the Netherlands and Germany), together with widespread natural seroconversion (post-infection), resulted in rapid eradication of both BTV-8 and BTV-1 from the region.
Although inactivated BTV vaccines were effective in northern Europe, some concerns still exist over the reliability of inactivation for each vaccine batch [23]. They are also monospecific, offering little protection against subsequent infections by heterologous BTV serotypes and it is uncertain how long the protective and neutralising antibodies responses that are generated will last in a vaccinated animal.
Although live attenuated vaccines are also available for multiple BTV serotypes, and appear to be highly effective in endemic regions for protection of individual susceptible animals against clinical signs of the disease, they can themselves cause severe disease in naïve sheep from northern Europe [24]. They also pose a further risk of genome segment re-assortment between vaccine and field strains, potentially leading to the emergence of progeny strains with novel biological characteristics.
Since both the inactivated and live attenuated BTV vaccines generate antibodies to all of the virus proteins, it has not been possible to develop a reliable serological assay to ‘distinguish infected from vaccinated animals’ (DIVA assays). In the face of a widespread vaccination campaign (as seen in Northern Europe) this invalidates the use of serological screening methods for surveillance. Virus detection, characterisation and surveillance during the BTV outbreaks in northern Europe have therefore relied heavily on conventional or real-time RT-PCR assays [25], [26].
Recombinant vaccinia virus, recombinant canarypox virus and recombinant capripox virus have all been used successfully as gene delivery systems for BTV-vaccination [27], . The passage of Chorioallantois vaccinia virus Ankara (CVA) over 570 times in primary chick embryo fibroblast cells (CEF), led to attenuated replication and reduced virulence, generating a new virus known as the ‘Modified Vaccinia Ankara’ strain (MVA) [30], [31]. Recombinant MVA (rMVA) provides a promising vaccine-vector, which activates both branches of the immune system [32], with a well-established safety record and history of use as a vaccine for infectious diseases and malignancies [31], [33]. DNA-vaccines have also been used experimentally for BTV and other orbiviruses [28], [34], [35], [36], [37] and were recently used in a heterologous prime-boost vaccination strategy (DNA/rMVA), providing protection against BTV in a mouse model system [28], [37].
Published data suggest that a combination of the three major BTV proteins VP2, VP5 and VP7, gives better protection than VP2 and VP5, or VP2 alone [5], [28], [38]. Indeed, co-expression of the four major structural proteins of BTV (VP2, VP3, VP5 and VP7 expressed by recombinant baculovirus) results in their assembly, generating ‘virus like particles’ (VLP) that also raise both neutralising antibodies and a protective response in sheep [38]. In addition, in vivo expression of the three capsid proteins VP2, VP5 and VP7 from three separate rMVA following a DNA prime/MVA boost vaccination regime was required to confer protective immunity in a BTV mouse model [28]. However, other studies with BTV and other related orbiviruses indicate that complete protection can be achieved by sub-unit vaccines based on the VP2 protein alone [39], [40]. In addition, an MVA based vaccine expressing VP2 of African horse sickness virus (AHSV) Serotype 4 also provided complete protection in mice against homologous AHSV-4 challenge [41].
The recent development of a new murine model, based on adult IFNAR (−/−) mice, has facilitated the study of the immune response and the testing of new vaccines against BTV. IFNAR (−/−) mice are knockout mice lacking the β subunit of the interferon α/β receptor and can be a good animal model for BTV because they are able to support the in vivo growth of BTV and they also show viraemia and disease symptoms [42]. Commercial inactivated vaccines against BTV have been tested in these mice [42] and they show very similar results comparing with vaccinated sheep or cattle in terms of neutralising antibodies and viraemia [43]. Moreover, our previous results [28], [37], [41], [42] and other studies [44], [45], [46], [47] show that the IFNAR (−/−) infection model is useful for the definition of effective recombinant vaccine candidates against several viruses.
In this study we investigated the protective efficacy of rMVA and DNA vaccines expressing BTV-8 VP2 in the mouse model based on IFNAR (−/−) mice. On the other hand, we wanted to determine whether the presence of VP5 and VP7 was critical in inducing protective immunity of VP2 based vaccines and whether a DNA prime/rMVA boost vaccination regime was more efficient than an rMVA prime/rMVA boost vaccination approach. We report the use of a heterologous DNA/rMVA and a homologous rMVA/rMVA prime-boost vaccine strategy of either BTV-8 VP2 (as sole antigen), or a combination of VP2, VP5 and VP7, to protect IFNAR (−/−) mice against a lethal challenge with a virulent strain of BTV-8.
Materials and Methods
Cells and Virus
Primary chicken embryo fibroblasts (CEF), the continuous chicken embryo fibroblast cell line DF-1 and African green monkey kidney cells (Vero) were all obtained from the Microbiological Services and Central Service units of the Institute for Animal Health.
The BTV-8 (Belgium/06 isolate) virus used for challenge studies at Centro de Investigación en Sanidad Animal, INIA, Madrid (CISA), was originally isolated from a calf in Belgium in 2006. The BTV-8 strain ‘NET2006/07′ from the orbivirus reference collection (ORC) at IAH (http://www.reoviridae.org/dsRNA_virus_proteins/ReoID/BTV-isolates.htm) was used for virus neutralisation tests (VNT) in Vero cells.
Generating DNA Vaccines
DNA vaccines were based on the pCI-neo Mammalian Expression Vector (Promega). To generate pCI-neo BTV-8 Seg-2, pCI-neo BTV-8 Seg-6 and pCI-neo BTV-8 Seg-7, we PCR amplified each BTV genome segment from plasmids pBRT7 BTV-8 Seg-2 NET2006/07, pBRT7 BTV-8 NET2006/07 Seg-6 and pBRT7 BTV-8 NET2006/07 Seg-7 using gene specific primers containing XbaI and NotI restriction sites (Table 1). The amplicons and recombinant pCI-neo were digested with XbaI and NotI and ligated using standard molecular cloning techniques and then sequenced using pCI-neo specific primers to identify the correct insert.
Table 1. Oligonucleotide primers.
| Name of Primer | Primer Sequence | Used to generate |
| BTV-8S2F/DNA | 5′-GCATTTTCTAGAATGGAGGAGCTAGCGATTCCGATTTAT-3′ | pCI-neo BTV-8 Seg-2 |
| BTV-8S2R/DNA | 5′-CGTAAAGCGGCCGCGCTATACATTGAGCAGCTTAGTTAACAT-3′ | pCI-neo BTV-8 Seg-2 |
| BTV-8S6F/DNA | 5′-GCATTTTCTAGAATGGGGAAAATCATAAAGTCC-3′ | pCI-neo BTV-8 Seg-6 |
| BTV-8S6R/DNA | 5′-AAATGCGCGGCCGCGTCAGGCATTTCTTAAGAAGAG-3′ | pCI-neo BTV-8 Seg-6 |
| BTV-8S7F/DNA | 5′-GCATTTTCTAGAATGGACACTATCGCTGCAAGAGCA-3′ | pCI-neo BTV-8 Seg-7 |
| BTV-8S7R/DNA | 5′-CGTAAAGCGGCCGCGCTAAGAGACGTTTGAATGGGTTAC-3′ | pCI-neo BTV-8 Seg-7 |
| BTV-8VP2F/VAC | 5′-TTTTCCCGGGACCATGGAGGAGCTAGCGATTCCGAT-3′ | pSC-11 BTV-8 Seg-2 |
| BTV8VP2R/VAC | 5′-TTTTCCCGGGCTATACATTGAGCAGCTTAG-3′ | pSC-11 BTV-8 Seg-2 |
| BTV-8VP5F/VAC | 5′-TTTTCCCGGGACCATGGGGAAAATCATAAAGTCCCTAAG-3′ | pSC-11 BTV-8 Seg-6 |
| BTV-8VP5R/VAC | 5′-TTTTCCCGGGTCAGGCATTTCTTAAGAAGAGTGG-3′ | pSC-11 BTV-8 Seg-6 |
| BTV-6VP7F/VAC | 5′-TTTTCCCGGGACCATGGACACTATCGCTGCAAGAGCAC-3′ | pSC-11 BTV-6 Seg-7 |
| BTV-6VP7R/VAC | 5′-TTTTCCCGGGCTAAGAGACGTTTGAATGGGTT-3′ | pSC-11 BTV-6 Seg-7 |
XbaI (TCTAGA), SmaI (CCCGGG) and NotI (GCGGCCGC) restriction sequences are underlined.
Generating Recombinant MVAs
Generation of rMVA BTV-8 VP2, rMVA BTV-8 VP5 and rMVA BTV-6 VP7 was done following standard methods [28]. Seg-7 was cloned from BTV-6 due to problems in cloning Seg-7 from BTV-8. However, it is a highly conserved segment used to identify the two phylogenetic groups [9] and was used in generating rMVA since blast analysis suggests BTV-6 Seg-7 and BTV-8 Seg-7 shares 95% amino acid identity. Briefly, the genes of interest were amplified from pBRT7 BTV-8 Seg-2 NET2006/07, pBRT7 BTV-8 NET2006/07 Seg-6 and pBRT7 BTV-6 NET2006/07 Seg-7 by PCR using gene specific primers (Table 1) containing a SmaI restriction site and cloned into the SmaI site of the standard vaccinia transfer vector pSC-11, downstream of the P7.5 vaccinia promoter, generating plasmids pSC-11 BTV-8 Seg- 2, pSC-11 BTV-8 Seg- 6 and pSC-11 BTV-6 Seg-7, respectively. DF-1 cells infected with MVA at an MOI of 0.1 were transfected with these recombinant plasmids using Lipofectamine™ 2000 Transfection Reagent (Invitrogen), to insert the pSC11 expression cassettes into the thymidine kinase gene locus of the MVA genome by homologous recombination. Recombinant viruses were selected by picking blue plaques following staining with X-gal and amplified in DF-1 cells. Transcription of BTV genes was checked by RT-PCR using specific primers (Table 1) and protein expression checked as detailed below.
Total RNA Extraction and RT-PCR
CEF were infected with rMVA BTV-8VP2, rMVA BTV-8 VP5 or rMVA BTV-6 VP7. At 24 hours post infection, the infected cells were harvested and centrifuged at 3000 rpm for 5 min. RNA was extracted from the pellet using RNeasy (Qiagen). RT-PCR was performed using Transcriptor One- Step RT-PCR kit (Roche) using primers BTV-8VP2F/VAC, BTV-8VP2R/VAC, BTV-8VP5F/VAC, BTV-8VP5R/VAC, BTV6-VP7F/VAC, and BTV-6VP7R/VAC (Table 1). Wild type MVA was used as negative control.
Detection of VP2, VP5 and VP7 Expressed by Recombinant MVAs or pCI-neo Plasmids by Indirect Immunofluorescence Assay
Detection of BTV proteins expressed by rMVAs or pCI-neo plasmids was carried out by indirect immunofluorescence assay. CEF cells were grown on cover slips and infected with rMVAs at a MOI of 0.1. On the other hand, Vero cells were transfected with the BTV pCI-neo plasmids using Lipofectamine 2000 (Invitrogen). After 24 h, cells were fixed with 4% paraformaldehyde for 30 min at room temperature. Permeabilization was performed with 0.4% Triton for 15 minutes before incubation with a PBS-20%FBS for 1 hour at room temperature. The cells were reacted with a serum from sheep infected with BTV-8 diluted 1∶800 in PBS-2%FBS for 2 h at 37°C, following washing with PBS. Alexa 488 conjugated polyclonal donkey anti-sheep IgG (Invitrogen) diluted 1∶1000 was used for fluorescent studies. Cells were washed and incubated for 5 minutes with DAPI. Finally cover slips were washed, mounted on glass slides and observed by confocal microscopy.
Immunisation of IFNAR (−/−) Mice and BTV-8 Challenge
Thirty six female IFNAR (−/−) mice were purchased from B&K Universal Ltd., United Kingdom. All experiments were performed under the guidelines of the European Community (86/609) and were approved by the ethical review committee (reference number: 2008/007) at the Centro de Investigación en Sanidad Animal, INIA, Madrid (CISA). Mice were maintained under pathogen-free conditions and allowed to acclimatize to biosafety level 3 (BSL3) animal facilities at CISA for 1 week before use.
Mice were divided in six groups for the experiment, five groups were immunised (three weeks apart) with the different vaccines while the remaining control group was not vaccinated (Table 2). A suspension of 100 µg of each DNA construct was administered intramuscularly. Doses of 3×107 pfu of rMVA-VP2 or 3×105 pfu of rMVA-VP5 or rMVA-VP7 were inoculated intraperitoneally, since it is the most employed route for MVA inoculation and has the additional advantage of the easier manipulation of animals. The dose used of rMVA-VP5 and rMVA-VP7 was lower because we could not obtain a higher titre of these viruses.
Table 2. Vaccination groups, dosage, route and schedule.
| Group | Vaccine | Dosage per mouse | Time |
| 1 | rMVA-VP2 prime | 3×107 pfu | Day 0 |
| 1 | rMVA-VP2 boost | 3×107 pfu | Day 21 |
| 2 | DNA-VP2 prime | 100 ìg | Day 0 |
| 2 | rMVA-VP2 boost | 3×107 pfu | Day 21 |
| 3 | DNA-VP7 prime | 100 ìg | Day 0 |
| 3 | rMVA-VP7 boost | 3×105 pfu | Day 21 |
| 4 | DNA-VP2, DNA-VP5, DNA-VP7 prime | 100 ìg each plasmid | Day 0 |
| 4 | MVA-VP2, MVA-VP5, MVA-VP7 boost | 3×107 pfu, 3×105 pfu, 3×105 pfu each | Day 21 |
| 5 | MVA-VP2, MVA-VP5, MVA-VP7prime | 3×107 pfu, 3×105 pfu, 3×105 pfu each | Day 0 |
| 5 | MVA-VP2, MVA-VP5, MVA-VP7 boost | 3×107 pfu, 3×105 pfu, 3×105 pfu each | Day 21 |
| 6 | Control | No vaccine | Day 0 |
| 6 | Control | No vaccine | Day 21 |
pfu = plaque forming units.
All of the mice were challenged two weeks after the second immunisation, using a lethal dose (10 pfu) of BTV-8 (Belgium/06 isolate) subcutaneously [42]. The clinical signs in vaccinated and control mice were monitored for 13 days post challenge and recorded. Animals that showed severe clinical signs (loss of more than 20% of body weight, frequent hunching, severe conjunctivitis or any other condition that prevented food or water intake) were humanely euthanized.
Blood Sampling, Virus Detection and Serology
All mice were bled using standard methods [48]. For serological analyses, blood samples were collected on days 0 (before prime immunisation), 20 and 34 post-vaccination and on day 7 and 13 post-challenge. Samples were incubated at room temperature for 30 minutes then centrifuged at 3000 rpm for 10 minutes.
The serum was collected and used for virus neutralisation test (VNT) as described previously [49]. Titres were assigned arithmetically as the dilution of serum that gave a 50% neutralisation endpoint and expressed as log10 values.
To determine viraemia titres, a standard plaque assay was conducted on EDTA blood samples collected on days 3, 5, 7, 10 and 12 pc. Whole blood samples were also analysed by a RT-qPCR assay specific for BTV segment 1 as previously described [25].
Statistical Methods
Differences amongst vaccine groups in outcome following challenge (i.e. survived or dead) were examined using a Fisher exact test. Differences in other measures (clinical score, onset of clinical signs, virus neutralising antibody titres, viral RNA levels and peak viral load) were examined using Kruskal-Wallis tests. If the Kruskal-Wallis test identified significant (P<0.05) differences amongst vaccine groups, these were explored in more detail using Wilcoxon tests for pairwise comparison between groups. Non parametric tests were preferred because of the small group sizes and potential non-normality of the errors.
Results
Expression of BTV VP2, VP5 and VP7 from rMVA and DNA Plasmid Vaccines
To test the functionality of the foreign gene expression cassettes in rMVA viruses and pCI-neo plasmids we performed RT-PCR amplification of VP2, VP5 and VP7 from total RNA extracted from rMVA infected or pCIneo transfected cells. We detected VP2, VP5 and VP7 specific cDNA amplicon bands of the expected sizes (∼ 2887, 1580 and 949 nucleotides for VP2, VP5 and VP7 respectively). Standard PCR using HotStart KoD DNA polymerase (Roche) showed no BTV specific DNA bands indicating the amplicons derived from BTV VP2, VP5 or VP7 RNA transcripts and not from plasmid (data not shown).
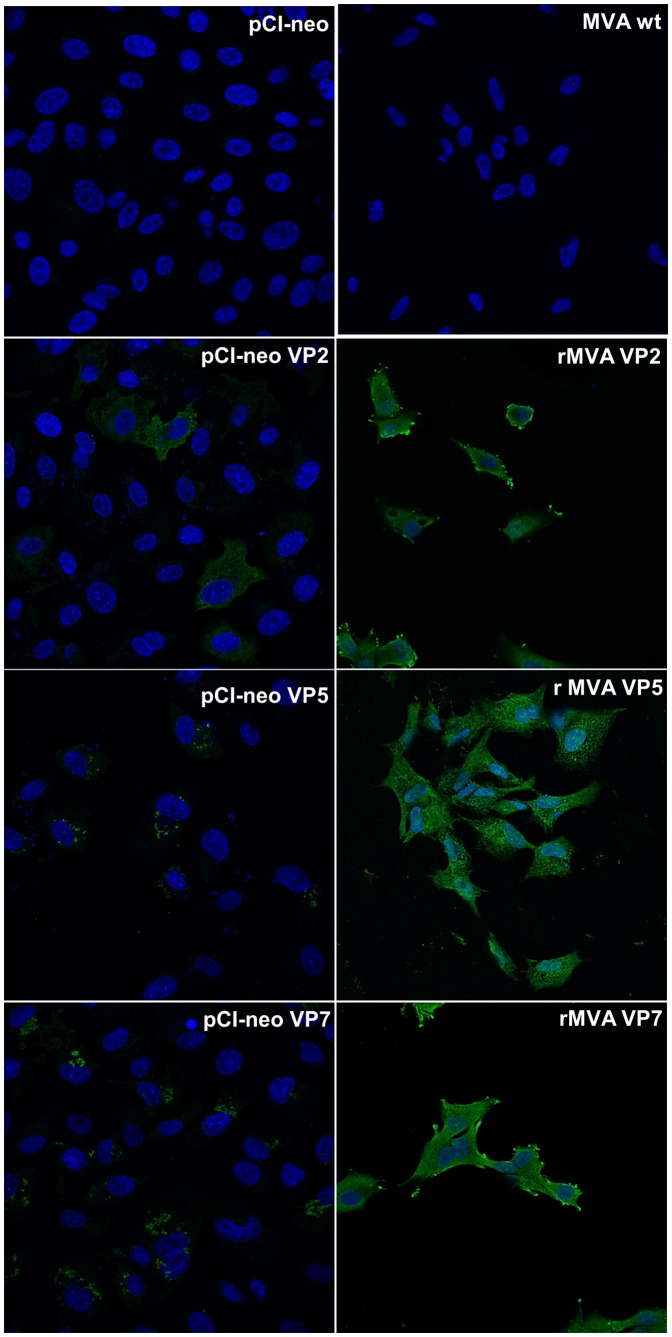
Moreover, we evaluated the expression of recombinant VP2, VP5 and VP7 by rMVAs or pCI-neo plasmids by immunofluorescence assays using a sheep anti BTV-8 serum (Fig. 1). Expression of these three proteins was confirmed in cells transfected with pCI-neo plasmids or infected with the rMVA viruses used for immunization of IFNAR (−/−) mice.
Figure 1. Expression of recombinant BTV proteins from rMVAs and pCi-neo plasmids by immunofluorescence.
CEF cells infected with rMVA-VP2, VP5 or VP7, or Vero cells transfected with pCI-neo VP2, VP5 or VP7 were analysis by immunofluorescence assay. Empty MVA and pCI-neo were used as negative controls. Fluorescence was observed on cells using a sheep serum anti BTV-8 followed by Alexa Fluor 488-conjugated donkey anti-sheep IgG. Nuclei were staining with DAPI. BTV protein expression from pCI-neo plasmids or rMVAs encoding VP2, VP5 and VP7 proteins was observed by confocal microscopy.
Post-challenge Clinical Signs
Mice from groups 1, 2, 4 and 5, vaccinated with VP2 (as sole antigen), or a combination of VP2, VP5 and VP7, using either a homologous (rMVA/rMVA) or a heterologous prime-boost (DNA/rMVA) vaccination regime, showed no clinical signs after challenge with BTV-8 and all of them survived (Table 3). Group 3 (vaccinated with heterologous DNA/rMVA expressing VP7 alone) and group 6 (control) showed severe clinical signs and were euthanized. However, in the VP7 vaccinated mice the onset of clinical signs was significantly delayed (Wilcoxon test: P = 0.01) in comparison with the control group. One animal of group 5 died immediately after bleeding without showing clinical signs or viraemia by plaque assay; therefore we considered that the death was not related with the infection.
Table 3. Clinical signs after challenge.
| Group | Mouse | Vaccine | Clinicalscore | Onset | Survival |
| 1 | 1.1 | MVA/MVA VP2 | 0 | Yes | |
| 1 | 1.2 | MVA/MVA VP2 | 0 | Yes | |
| 1 | 1.3 | MVA/MVA VP2 | 0 | Yes | |
| 1 | 1.4 | MVA/MVA VP2 | 0 | Yes | |
| 1 | 1.5 | MVA/MVA VP2 | 0 | Yes | |
| 1 | 1.6 | MVA/MVA VP2 | 0 | Yes | |
| 2 | 2.1 | DNA/MVA VP2 | 0 | Yes | |
| 2 | 2.2 | DNA/MVA VP2 | 0 | Yes | |
| 2 | 2.3 | DNA/MVA VP2 | 0 | Yes | |
| 2 | 2.4 | DNA/MVA VP2 | 0 | Yes | |
| 2 | 2.5 | DNA/MVA VP2 | 0 | Yes | |
| 2 | 2.6 | DNA/MVA VP2 | 0 | Yes | |
| 3 | 3.1 | DNA/MVA VP7 | 3 | Day 5 | No |
| 3 | 3.2 | DNA/MVA VP7 | 4 | Day 5 | No |
| 3 | 3.3 | DNA/MVA VP7 | 4 | Day 5 | No |
| 3 | 3.4 | DNA/MVA VP7 | 4 | Day 5 | No |
| 3 | 3.5 | DNA/MVA VP7 | 3 | Day 6 | No |
| 3 | 3.6 | DNA/MVA VP7 | 6 | Day 4 | No |
| 4 | 4.1 | DNA/MVA VP2 VP5 VP7 | 0 | Yes | |
| 4 | 4.2 | DNA/MVA VP2 VP5 VP7 | 0 | Yes | |
| 4 | 4.3 | DNA/MVA VP2 VP5 VP7 | 0 | Yes | |
| 4 | 4.4 | DNA/MVA VP2 VP5 VP7 | 0 | Yes | |
| 4 | 4.5 | DNA/MVA VP2 VP5 VP7 | 0 | Yes | |
| 4 | 4.6 | DNA/MVA VP2 VP5 VP7 | 0 | Yes | |
| 5 | 5.1 | MVA/MVA VP2 VP5 VP7 | 0 | Yes | |
| 5 | 5.2 | MVA/MVA VP2 VP5 VP7 | 0 | Yes | |
| 5 | 5.3 | MVA/MVA VP2 VP5 VP7 | 0 | Yes | |
| 5 | 5.4 | MVA/MVA VP2 VP5 VP7 | 0 | Yes | |
| 5 | 5.5 | MVA/MVA VP2 VP5 VP7 | 0 | Yes | |
| 5 | 5.6 | MVA/MVA VP2 VP5 VP7 | 0 | ||
| 6 | 6.1 | Control | 4 | Day 4 | No |
| 6 | 6.2 | Control | 4 | Day 4 | No |
| 6 | 6.3 | Control | 6 | Day 3 | No |
| 6 | 6.4 | Control | 5 | Day 4 | No |
| 6 | 6.5 | Control | 4 | Day 4 | No |
| 6 | 6.6 | Control | 3 | Day 4 | No |
Clinical signs from individual mice were recorded and assigned a value according to the following algorithm: reduced activity: 1; frequent hunching: 2; ruffled fur: 1; weight loss: 2; swelling around the eyes: 1. The final clinical score for each animal was the sum of all the values for each individual. Day of onset is the day after challenge when clinical signs appeared.
Viraemia in Mice after Challenge with BTV-8
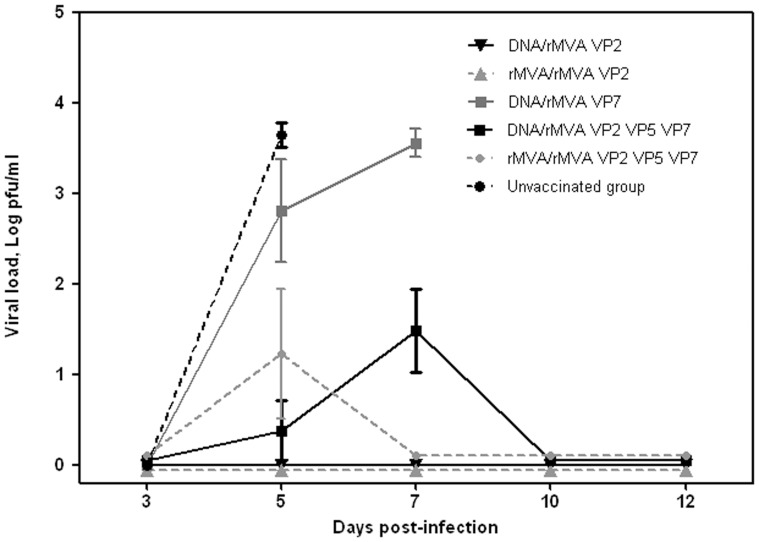
No virus was detected in any blood samples taken on day 3 pc. All mice in groups 3 and 6 (vaccinated with VP7 and unvaccinated controls, respectively) developed viraemia titres higher than 1×103 pfu/ml prior to death (Figure 2). Three mice in group 4 and two mice in group 5 (both groups vaccinated with VP2, VP5 and VP7, which all survived) also developed viraemia, albeit at significantly (P = 0,002) lower titres than in groups 3 and 6 (≤3×102 pfu/ml). Although mice in groups 1, 2, 4 and 5 were protected and survived until the end of the experiment, only mice vaccinated with VP2 (Groups 1 and 2) showed no viraemia by plaque assay.
Figure 2. Viral load (log pfu/ml) of blood samples collected from vaccinated and control mice after challenge with BTV-8.
Virus was extracted from blood of immunized and non-immunized mice after challenge and determined as described in Materials and Methods. Each point represents the mean values of the viral titer of six animals and standard errors are shown as bars.
Detection of Viral RNA in Blood from Mice Post Challenge
The results summarised on Table 4 show that five control mice (Group 6) were positive on day 3 and all six animals were positive on day 5 pc, prior to death. Group 3 (vaccinated with VP7 alone) showed a similar pattern, with three mice becoming positive on day 3. All mice in group 3 were positive on day 5 and the two that survived beyond day 7 were positive on that day. In groups 4 and 5, BTV RNA was detected throughout the experiment, from day 3 to day 12. In Group 1, BTV RNA was detected in five mice but only on day 7 pc; whilst in group 2, BTV RNA was only detected in a single mouse on days 5 and 7 pc. The observed Ct values on day 5 were significantly (P<0.02) higher for mice in groups 1 and 2 compared with those in groups 3, 4, 5 and 6 and for mice in groups 4 and 5 compared with those in group 6. In addition, the observed Ct values on day 7 were significantly (P = 0.05) higher in mice for groups 1 and 2 compared with those in group 4 and 5.
Table 4. BTV-8 RNA detection (Ct values) in blood samples collected from vaccinated and control mice after challenge.
| Group | Mouse | Vaccine | Day 3 | Day 5 | Day 7 | Day 10 | Day 12 |
| 1 | 1.1 | MVA/MVA VP2 | – | – | 34.7 | – | – |
| 1 | 1.2 | MVA/MVA VP2 | – | – | 33.72 | – | – |
| 1 | 1.3 | MVA/MVA VP2 | – | – | 34.24 | – | – |
| 1 | 1.4 | MVA/MVA VP2 | – | – | – | – | – |
| 1 | 1.5 | MVA/MVA VP2 | – | – | 33.93 | – | – |
| 1 | 1.6 | MVA/MVA VP2 | – | – | 34.3 | – | – |
| 2 | 2.1 | DNA/MVA VP2 | – | – | – | – | – |
| 2 | 2.2 | DNA/MVA VP2 | – | – | – | – | – |
| 2 | 2.3 | DNA/MVA VP2 | – | 33.24 | 32.95 | – | – |
| 2 | 2.4 | DNA/MVA VP2 | – | – | – | – | – |
| 2 | 2.5 | DNA/MVA VP2 | – | – | – | – | – |
| 2 | 2.6 | DNA/MVA VP2 | – | – | – | – | – |
| 3 | 3.1 | DNA/MVA VP7 | 35.15 | 26.34 | † | ||
| 3 | 3.2 | DNA/MVA VP7 | 33.86 | 24.67 | † | ||
| 3 | 3.3 | DNA/MVA VP7 | – | 30.67 | † | ||
| 3 | 3.4 | DNA/MVA VP7 | – | 32.33 | 25.6 | † | |
| 3 | 3.5 | DNA/MVA VP7 | – | 32.7 | 31.2 | † | |
| 3 | 3.6 | DNA/MVA VP7 | 31.79 | 23.85 | † | ||
| 4 | 4.1 | DNA/MVA VP2 VP5 VP7 | – | 34.9 | 31.55 | 31.37 | 32.8 |
| 4 | 4.2 | DNA/MVA VP2 VP5 VP7 | 40.35 | 32.51 | 30.37 | 34.62 | 33.64 |
| 4 | 4.3 | DNA/MVA VP2 VP5 VP7 | 33.91 | 31.47 | 29.96 | – | – |
| 4 | 4.4 | DNA/MVA VP2 VP5 VP7 | 34.2 | 31.55 | 30.7 | 31.97 | – |
| 4 | 4.5 | DNA/MVA VP2 VP5 VP7 | 31.9 | 31.47 | 31.9 | 31.33 | 33.62 |
| 4 | 4.6 | DNA/MVA VP2 VP5 VP7 | – | 33.65 | 35.07 | – | 34.2 |
| 5 | 5.1 | MVA/MVA VP2 VP5 VP7 | – | 30.42 | 31.16 | – | – |
| 5 | 5.2 | MVA/MVA VP2 VP5 VP7 | – | 31.1 | 31.32 | 32.57 | 33.03 |
| 5 | 5.3 | MVA/MVA VP2 VP5 VP7 | – | 35.04 | 33.84 | 32.44 | – |
| 5 | 5.4 | MVA/MVA VP2 VP5 VP7 | 34.13 | 42.06 | 32.61 | 30.85 | 34.77 |
| 5 | 5.5 | MVA/MVA VP2 VP5 VP7 | 33.85 | – | 34.49 | – | – |
| 5 | 5.6 | MVA/MVA VP2 VP5 VP7 | 34.39 | 30.47 | † | † | |
| 6 | 6.1 | Control | 30.34 | 25.94 | † | ||
| 6 | 6.2 | Control | 32.4 | 24.82 | † | ||
| 6 | 6.3 | Control | 31.38 | 24.07 | † | ||
| 6 | 6.4 | Control | 34.56 | 28.02 | † | ||
| 6 | 6.5 | Control | – | 29.09 | † | ||
| 6 | 6.6 | Control | 33.87 | 26.46 | † |
No Ct value for a particular sample is indicated by –.
Sample not taken due to previous death of the animal is indicated by †.
Neutralising Antibodies in Vaccinated Mice
Virus neutralising antibodies against BTV-8 were detected on day 34 (two weeks post boost) in all mice that had received VP2 based vaccines (DNA/rMVA or rMVA/rMVA), either alone or in combination with other BTV proteins (Groups 1, 2, 4, and 5) (Figure 3). Titres ranged between 1.06 and 1.15 (log10 VNT) and did not differ significantly (P>0.05) amongst these groups. Titres rose after challenge ranged from 1.88 to 3. No neutralising antibodies were detected in serum from mice vaccinated with VP7 alone, or in serum from the control group (Group 6). The level of neutralising antibodies post-challenge was significantly (P<0.02) higher in group 5 compared with groups 1, 2 and 4 possibly caused by the partial replication of the challenge viruses. In contrast, groups 1, 2 and 4, which developed slightly higher neutralising antibodies post vaccination, were more effectively protected, with lower levels of challenge-virus replication, and developed lower neutralising antibody levels by day 13 pc.
Figure 3. Virus neutralising antibodies in serum of mice following vaccination and challenge.
Mean values of AHSV-4 neutralising antibodies measured in mice groups at different times after vaccination and after challenge. Titres are assigned arithmetically as the dilution of serum that will give a 50% neutralisation endpoint and expressed as log10 values. Standard deviations are shown as error bars. p.v. = post-vaccination, p.i. = post-infection.
Discussion
Classical vaccines have been used against BTV in the field, however recombinant vaccines offer good immunogenicity and are safer avoiding reversion to virulence (e.g. by reassortment with wild type strains), contamination with toxic compounds used for inactivation and any risk of incomplete inactivation of whole-cell vaccines. Moreover, recombinant marker vaccines allow the distinction between vaccinated and naturally infected animals (DIVA). Recombinant live viruses and DNA plasmids have been widely used as delivery systems for expression of foreign antigens and as vaccine candidates for infectious diseases and cancer [50], [51], [52], [53]. DNA vaccines appear to be more efficacious when they are used to prime immune responses in heterologous vaccination regimes with recombinant Adenovirus or Poxvirus vectors. Indeed, it has been described that these heterologous protocols induced higher frequencies of CTLs than when each immunogen was administered separately [54], [55]. However, homologous prime-boost vaccination with recombinant MVA virus (Family Poxviridae) has been used in many successful vaccination studies against hepatitis C [56], HIV [57] and influenza A/H5N1 [58] viruses, demonstrating its efficacy as potential vaccine following that regime. Indeed, it has been described that recombinant MVA vaccines can be administered repeatedly without interference of vector-specific antibodies induced after the first immunization and without loss of booster antibody responses against the target antigen after subsequent immunization [59], [60].
In the case of BTV, previous results of DNA/rMVA prime-boost vaccination using VP2, VP5 and VP7 from BTV-4 in a mouse model [29] demonstrated the efficacy of this approach. On the other hand, in earlier studies with AHSV [41], [61] we also observed that rMVA expressing AHSV-4 VP2, when used in a homologous prime-boost vaccination regime, induced good neutralizing antibody responses in ponies and neutralizing antibodies and protection in IFNAR (−/−) mice. In this study, we have compared directly the efficacy of homologous rMVA/rMVA and heterologous DNA/rMVA prime-boost vaccination regimes expressing either VP2 alone, VP7 alone or a combination of VP2, VP5 and VP7 proteins in IFNAR (−/−) mice.
Our results showed that both homologous (rMVA/rMVA) or heterologous (DNA/rMVA) prime boost vaccinations, expressing either BTV-8 VP2 alone or a combination of three major BTV proteins VP2, VP5 and VP7, induced protective immunity against BTV-8 challenge in mice. We showed very similar levels of efficacy of both vaccination regimes. The two groups of mice vaccinated with BTV-8 VP2 alone, using either a heterologous DNA/rMVA or homologous rMVA/rMVA strategy, were completely protected against clinical signs of BTV infection and had no detectable viraemia by plaque assay. Only low level of BTV RNA was detected in some individuals by qRT-PCR.
The present study also showed that the addition of DNAs or MVAs expressing VP5 and VP7 was not critical for the induction of neutralising antibodies and protection. These vaccines did not improve the protection induced by BTV-8 VP2 alone following DNA/rMVA or rMVA/rMVA vaccinations, since vaccination with BTV-8 VP2 alone was enough to protect animals. Indeed, mice immunized with a combination of recombinant vaccines each expressing VP2, VP5 or VP7 showed higher levels of BTV virus and BTV RNA in blood than mice immunised with VP2 alone. This result contrasts with those observed in previous studies with MVA BTV-4 [28], where VP2, VP5 and VP7 were necessary to confer protective immunity in IFNAR (−/−) mice. In our present study the protein VP2 was expressed from BTV-8, indicating that there could be differences in immunogenicity between same proteins of different serotypes. Moreover, VP2 isolated from BTV virus particles or as expressed by recombinant baculoviruses, has previously been used to protect sheep from BTV challenge [13], [38], [62]. In other studies BTV-8 VP2 alone was not enough to confer protection against challenge in mice, nevertheless the viral vectors used in those studies were different from MVA [49], [63]. At present it is not clear why in some circumstances VP2 alone is enough to induce protective immunity. Further studies would be necessary to better characterise the induction of immune responses following these vaccinations.
Although VP7 does not raise antibodies that can neutralise intact BTV particles, it can provide partial protection via a cell mediated immune response, and its incorporation is thought to enhance the efficacy of VP2 and VP5 vaccines [38], [64]. In the current study, although vaccination with VP7 alone did not protect IFNAR (−/−) mice against BTV-8 challenge, animals showed a delayed onset of clinical signs and the survival time was slightly longer than in the non-vaccinated mice.
In summary, our results show that VP2 expressed in vivo using a heterologous or homologous prime boost vaccination (DNA/rMVA or rMVA/rMVA), can generate immunity against BTV-8 in IFNAR (−/−) mice, protecting them against a lethal challenge, and that a homologous vaccination regime using rMVA was at least as effective as a DNA/rMVA heterologous approach. However, further work will be needed to test and validate the use and efficacy of these BTV-subunit vaccine candidates in ruminants, the natural hosts for BTV infection.
Acknowledgments
The authors are grateful to Andrew Shaw for providing pBRT7 BTV-8 Seg-2 NET2006/07, pBRT7 BTV-8 NET2006/07 Seg-6, pBRT7 BTV-8 NET2006/07 Seg-7 and pBRT7 BTV-6 NET2006/07 Seg-7.
Funding Statement
This work was supported by Defra, BBSRC, Wellcome Trust, the European Commission and the Comisión Interministerial de Ciencia y Tecnología (CICYT) (OrbiVac - Grant no.: 245266; WildTech - Grant no.: 222633-2; EMIDA grant OrbiNet - K1303206; and grant AGL2011-23506/GAN). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Erasmus BJ (1975) Bluetongue in sheep and goats. Aust Vet J 51: 165–170. [DOI] [PubMed] [Google Scholar]
- 2. Darpel KE, Batten CA, Veronesi E, Shaw AE, Anthony S, et al. (2007) Clinical signs and pathology shown by British sheep and cattle infected with bluetongue virus serotype 8 derived from the 2006 outbreak in northern Europe. Vet Rec 161: 253–261. [DOI] [PubMed] [Google Scholar]
- 3. Mertens PP, Brown F, Sangar DV (1984) Assignment of the genome segments of bluetongue virus type 1 to the proteins which they encode. Virology 135: 207–217. [DOI] [PubMed] [Google Scholar]
- 4. Belhouchet M, Mohd Jaafar F, Firth AE, Grimes JM, Mertens PP, et al. (2011) Detection of a fourth orbivirus non-structural protein. PLoS One 6: e25697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Roy P (1992) From genes to complex structures of bluetongue virus and their efficacy as vaccines. Vet Microbiol 33: 155–168. [DOI] [PubMed] [Google Scholar]
- 6. Roy P (1992) Bluetongue virus proteins. J Gen Virol 73 (Pt 12): 3051–3064. [DOI] [PubMed] [Google Scholar]
- 7. Grimes JM, Burroughs JN, Gouet P, Diprose JM, Malby R, et al. (1998) The atomic structure of the bluetongue virus core. Nature 395: 470–478. [DOI] [PubMed] [Google Scholar]
- 8. Bonneau KR, Mullens BA, MacLachlan NJ (2001) Occurrence of genetic drift and founder effect during quasispecies evolution of the VP2 and NS3/NS3A genes of bluetongue virus upon passage between sheep, cattle, and Culicoides sonorensis. J Virol 75: 8298–8305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Anthony S, Jones H, Darpel KE, Elliott H, Maan S, et al. (2007) A duplex RT-PCR assay for detection of genome segment 7 (VP7 gene) from 24 BTV serotypes. J Virol Methods 141: 188–197. [DOI] [PubMed] [Google Scholar]
- 10. Maan S, Maan NS, Samuel AR, Rao S, Attoui H, et al. (2007) Analysis and phylogenetic comparisons of full-length VP2 genes of the 24 bluetongue virus serotypes. J Gen Virol 88: 621–630. [DOI] [PubMed] [Google Scholar]
- 11. Maan S, Maan NS, Ross-smith N, Batten CA, Shaw AE, et al. (2008) Sequence analysis of bluetongue virus serotype 8 from the Netherlands 2006 and comparison to other European strains. Virology 377: 308–318. [DOI] [PubMed] [Google Scholar]
- 12. Maan S, Maan NS, Nomikou K, Veronesi E, Bachanek-Bankowska K, et al. (2011) Complete genome characterisation of a novel 26th bluetongue virus serotype from Kuwait. PLoS One 6: e26147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Huismans H, van der Walt NT, Cloete M, Erasmus BJ (1987) Isolation of a capsid protein of bluetongue virus that induces a protective immune response in sheep. Virology 157: 172–179. [DOI] [PubMed] [Google Scholar]
- 14. DeMaula CD, Bonneau KR, MacLachlan NJ (2000) Changes in the outer capsid proteins of bluetongue virus serotype ten that abrogate neutralization by monoclonal antibodies. Virus Res 67: 59–66. [DOI] [PubMed] [Google Scholar]
- 15. Mertens PP, Pedley S, Cowley J, Burroughs JN, Corteyn AH, et al. (1989) Analysis of the roles of bluetongue virus outer capsid proteins VP2 and VP5 in determination of virus serotype. Virology 170: 561–565. [DOI] [PubMed] [Google Scholar]
- 16. Cowley JA, Gorman BM (1987) Genetic reassortants for identification of the genome segment coding for the bluetongue virus hemagglutinin. J Virol 61: 2304–2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hutchinson I (1999) The role of VP7 in initiation of infection by bluetongue virus. United Kingdom: University of Hertfordshire.
- 18. Mertens PP, Burroughs JN, Walton A, Wellby MP, Fu H, et al. (1996) Enhanced infectivity of modified bluetongue virus particles for two insect cell lines and for two Culicoides vector species. Virology 217: 582–593. [DOI] [PubMed] [Google Scholar]
- 19. Mertens PP, Maan NS, Prasad G, Samuel AR, Shaw AE, et al. (2007) Design of primers and use of RT-PCR assays for typing European bluetongue virus isolates: differentiation of field and vaccine strains. J Gen Virol 88: 2811–2823. [DOI] [PubMed] [Google Scholar]
- 20. Hofmann M, Griot C, Chaignat V, Perler L, Thur B (2008) Bluetongue disease reaches Switzerland. Schweiz Arch Tierheilkd 150: 49–56. [DOI] [PubMed] [Google Scholar]
- 21. Saegerman C, Mellor P, Uyttenhoef A, Hanon JB, Kirschvink N, et al. (2010) The most likely time and place of introduction of BTV8 into Belgian ruminants. PLoS One 5: e9405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gloster J, Burgin L, Witham C, Athanassiadou M, Mellor PS (2008) Bluetongue in the United Kingdom and northern Europe in 2007 and key issues for 2008. Vet Rec 162: 298–302. [DOI] [PubMed] [Google Scholar]
- 23. Gethmann J, Huttner K, Heyne H, Probst C, Ziller M, et al. (2009) Comparative safety study of three inactivated BTV-8 vaccines in sheep and cattle under field conditions. Vaccine 27: 4118–4126. [DOI] [PubMed] [Google Scholar]
- 24. Veronesi E, Darpel KE, Hamblin C, Carpenter S, Takamatsu HH, et al. (2010) Viraemia and clinical disease in Dorset Poll sheep following vaccination with live attenuated bluetongue virus vaccines serotypes 16 and 4. Vaccine 28: 1397–1403. [DOI] [PubMed] [Google Scholar]
- 25. Shaw AE, Monaghan P, Alpar HO, Anthony S, Darpel KE, et al. (2007) Development and initial evaluation of a real-time RT-PCR assay to detect bluetongue virus genome segment 1. J Virol Methods 145: 115–126. [DOI] [PubMed] [Google Scholar]
- 26. Elia G, Savini G, Decaro N, Martella V, Teodori L, et al. (2008) Use of real-time RT-PCR as a rapid molecular approach for differentiation of field and vaccine strains of bluetongue virus serotypes 2 and 9. Mol Cell Probes 22: 38–46. [DOI] [PubMed] [Google Scholar]
- 27. Boone JD, Balasuriya UB, Karaca K, Audonnet JC, Yao J, et al. (2007) Recombinant canarypox virus vaccine co-expressing genes encoding the VP2 and VP5 outer capsid proteins of bluetongue virus induces high level protection in sheep. Vaccine 25: 672–678. [DOI] [PubMed] [Google Scholar]
- 28. Calvo-Pinilla E, Rodriguez-Calvo T, Sevilla N, Ortego J (2009) Heterologous prime boost vaccination with DNA and recombinant modified vaccinia virus Ankara protects IFNAR(−/−) mice against lethal bluetongue infection. Vaccine 28: 437–445. [DOI] [PubMed] [Google Scholar]
- 29. Savini G, MacLachlan NJ, Sanchez-Vizcaino JM, Zientara S (2008) Vaccines against bluetongue in Europe. Comp Immunol Microbiol Infect Dis 31: 101–120. [DOI] [PubMed] [Google Scholar]
- 30. Mayr A, Stickl H, Muller HK, Danner K, Singer H (1978) The smallpox vaccination strain MVA: marker, genetic structure, experience gained with the parenteral vaccination and behavior in organisms with a debilitated defence mechanism (author’s transl). Zentralbl Bakteriol B 167: 375–390. [PubMed] [Google Scholar]
- 31. Esteban M (2009) Attenuated poxvirus vectors MVA and NYVAC as promising vaccine candidates against HIV/AIDS. Hum Vaccin 5: 867–871. [DOI] [PubMed] [Google Scholar]
- 32. Ramirez JC, Gherardi MM, Esteban M (2000) Biology of attenuated modified vaccinia virus Ankara recombinant vector in mice: virus fate and activation of B- and T-cell immune responses in comparison with the Western Reserve strain and advantages as a vaccine. J Virol 74: 923–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kennedy JS, Greenberg RN (2009) IMVAMUNE: modified vaccinia Ankara strain as an attenuated smallpox vaccine. Expert Rev Vaccines 8: 13–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Huismans H (1985) The use of recombinant DNA technology for the development of a bluetongue virus subunit vaccine. Onderstepoort J Vet Res 52: 149–151. [PubMed] [Google Scholar]
- 35. Romito M, Du Plessis DH, Viljoen GJ (1999) Immune responses in a horse inoculated with the VP2 gene of African horsesickness virus. Onderstepoort J Vet Res 66: 139–144. [PubMed] [Google Scholar]
- 36. MacLachlan NJ, Balasuriya UB, Davis NL, Collier M, Johnston RE, et al. (2007) Experiences with new generation vaccines against equine viral arteritis, West Nile disease and African horse sickness. Vaccine 25: 5577–5582. [DOI] [PubMed] [Google Scholar]
- 37. Calvo-Pinilla E, Navasa N, Anguita J, Ortego J (2012) Multiserotype protection elicited by a combinatorial prime-boost vaccination strategy against bluetongue virus. PLoS One 7: e34735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Roy P (1990) Use of baculovirus expression vectors: development of diagnostic reagents, vaccines and morphological counterparts of bluetongue virus. FEMS Microbiol Immunol 2: 223–234. [DOI] [PubMed] [Google Scholar]
- 39. Scanlen M, Paweska JT, Verschoor JA, van Dijk AA (2002) The protective efficacy of a recombinant VP2-based African horsesickness subunit vaccine candidate is determined by adjuvant. Vaccine 20: 1079–1088. [DOI] [PubMed] [Google Scholar]
- 40. Stone-Marschat MA, Moss SR, Burrage TG, Barber ML, Roy P, et al. (1996) Immunization with VP2 is sufficient for protection against lethal challenge with African horsesickness virus Type 4. Virology 220: 219–222. [DOI] [PubMed] [Google Scholar]
- 41. Castillo-Olivares J, Calvo-Pinilla E, Casanova I, Bachanek-Bankowska K, Chiam R, et al. (2011) A modified vaccinia Ankara virus (MVA) vaccine expressing African horse sickness virus (AHSV) VP2 protects against AHSV challenge in an IFNAR −/− mouse model. PLoS One 6: e16503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Calvo-Pinilla E, Rodriguez-Calvo T, Anguita J, Sevilla N, Ortego J (2009) Establishment of a bluetongue virus infection model in mice that are deficient in the alpha/beta interferon receptor. PLoS One 4: e5171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Paradell H, Garcia L, Urniza A, Vila A, San Miguel E, et al.. (2008) Efficacy of ZULVAC® inactivated and adjuvanted vaccine against bluetongue virus Bluetongue minisymposium (Brescia).
- 44. Fiette L, Aubert C, Muller U, Huang S, Aguet M, et al. (1995) Theiler’s virus infection of 129Sv mice that lack the interferon alpha/beta or interferon gamma receptors. J Exp Med 181: 2069–2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ohka S, Igarashi H, Nagata N, Sakai M, Koike S, et al. (2007) Establishment of a poliovirus oral infection system in human poliovirus receptor-expressing transgenic mice that are deficient in alpha/beta interferon receptor. J Virol 81: 7902–7912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Waibler Z, Anzaghe M, Ludwig H, Akira S, Weiss S, et al. (2007) Modified vaccinia virus Ankara induces Toll-like receptor-independent type I interferon responses. J Virol 81: 12102–12110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ma G, Eschbaumer M, Said A, Hoffmann B, Beer M, et al. (2012) An equine herpesvirus type 1 (EHV-1) expressing VP2 and VP5 of serotype 8 bluetongue virus (BTV-8) induces protection in a murine infection model. PLoS One 7: e34425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Golde WT, Gollobin P, Rodriguez LL (2005) A rapid, simple, and humane method for submandibular bleeding of mice using a lancet. Lab Anim (NY) 34: 39–43. [DOI] [PubMed] [Google Scholar]
- 49. Franceschi V, Capocefalo A, Calvo-Pinilla E, Redaelli M, Mucignat-Caretta C, et al. (2011) Immunization of knock-out alpha/beta interferon receptor mice against lethal bluetongue infection with a BoHV-4-based vector expressing BTV-8 VP2 antigen. Vaccine 29: 3074–3082. [DOI] [PubMed] [Google Scholar]
- 50. Wang Z, Martinez J, Zhou W, La Rosa C, Srivastava T, et al. (2010) Modified H5 promoter improves stability of insert genes while maintaining immunogenicity during extended passage of genetically engineered MVA vaccines. Vaccine 28: 1547–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Anderson RJ, Schneider J (2007) Plasmid DNA and viral vector-based vaccines for the treatment of cancer. Vaccine 25 Suppl 2B24–34. [DOI] [PubMed] [Google Scholar]
- 52. Dunham SP (2002) The application of nucleic acid vaccines in veterinary medicine. Res Vet Sci 73: 9–16. [DOI] [PubMed] [Google Scholar]
- 53. Moore AC, Hill AV (2004) Progress in DNA-based heterologous prime-boost immunization strategies for malaria. Immunol Rev 199: 126–143. [DOI] [PubMed] [Google Scholar]
- 54. Kent SJ, Zhao A, Best SJ, Chandler JD, Boyle DB, et al. (1998) Enhanced T-cell immunogenicity and protective efficacy of a human immunodeficiency virus type 1 vaccine regimen consisting of consecutive priming with DNA and boosting with recombinant fowlpox virus. J Virol 72: 10180–10188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Amara RR, Villinger F, Altman JD, Lydy SL, O’Neil SP, et al. (2001) Control of a mucosal challenge and prevention of AIDS by a multiprotein DNA/MVA vaccine. Science 292: 69–74. [DOI] [PubMed] [Google Scholar]
- 56. Fournillier A, Gerossier E, Evlashev A, Schmitt D, Simon B, et al. (2007) An accelerated vaccine schedule with a poly-antigenic hepatitis C virus MVA-based candidate vaccine induces potent, long lasting and in vivo cross-reactive T cell responses. Vaccine 25: 7339–7353. [DOI] [PubMed] [Google Scholar]
- 57. Earl PL, Cotter C, Moss B, VanCott T, Currier J, et al. (2009) Design and evaluation of multi-gene, multi-clade HIV-1 MVA vaccines. Vaccine 27: 5885–5895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kreijtz JH, Suezer Y, de Mutsert G, van den Brand JM, van Amerongen G, et al. (2009) Preclinical evaluation of a modified vaccinia virus Ankara (MVA)-based vaccine against influenza A/H5N1 viruses. Vaccine 27: 6296–6299. [DOI] [PubMed] [Google Scholar]
- 59. Harrop R, Connolly N, Redchenko I, Valle J, Saunders M, et al. (2006) Vaccination of colorectal cancer patients with modified vaccinia Ankara delivering the tumor antigen 5T4 (TroVax) induces immune responses which correlate with disease control: a phase I/II trial. Clin Cancer Res 12: 3416–3424. [DOI] [PubMed] [Google Scholar]
- 60. Ramirez JC, Gherardi MM, Rodriguez D, Esteban M (2000) Attenuated modified vaccinia virus Ankara can be used as an immunizing agent under conditions of preexisting immunity to the vector. J Virol 74: 7651–7655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Chiam R, Sharp E, Maan S, Rao S, Mertens P, et al. (2009) Induction of antibody responses to African horse sickness virus (AHSV) in ponies after vaccination with recombinant modified vaccinia Ankara (MVA). PLoS One 4: e5997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Roy P, Urakawa T, Van Dijk AA, Erasmus BJ (1990) Recombinant virus vaccine for bluetongue disease in sheep. J Virol 64: 1998–2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Ma G, Eschbaumer M, Said A, Hoffmann B, Beer M, et al. (2012) An equine herpesvirus type 1 (EHV-1) expressing VP2 and VP5 of serotype 8 bluetongue virus (BTV-8) induces protection in a murine infection model. PLoS One 7: e34425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wade-Evans AM, Woolhouse T, O’Hara R, Hamblin C (1993) The use of African horse sickness virus VP7 antigen, synthesised in bacteria, and anti-VP7 monoclonal antibodies in a competitive ELISA. J Virol Methods 45: 179–188. [DOI] [PubMed] [Google Scholar]