Figure 1. Rescue and characterization of rSeVhHN, rSeVh(HN+F) and rSeVh(HN+Fstail).
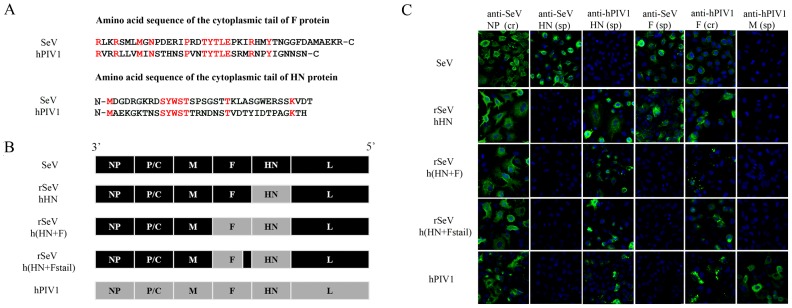
(A) Aligned amino acid sequences of the F and HN cytoplasmic tails of hPIV1 and SeV. Conserved amino acids are highlighted in red. (B) Schematic diagram of rSeV genomes compared with wt SeV and hPIV1. With rSeVhHN, the SeV HN gene was replaced with that of hPIV1. In rSeVh(HN+F), both HN and F genes of SeV were replaced with those of hPIV1. For rSeVh(HN+Fstail), the entire cytoplasmic tail portion of hPIV1 F shown in Figure 1A was replaced with that of SeV F. (C) Expression of HN and F from rSeVs. IF analysis of A549 cells infected with wt SeV, hPIV1 or rSeV. Origins of HN or F were confirmed using cross reactive (cr) or specific (sp) mAb for NP (cr M52), HN (SeV-sp S16, hPIV1-sp P24), F (SeV-sp M38, cr P38) or M (hPIV1-sp P3) [12], [36], [41], [42], [43].
