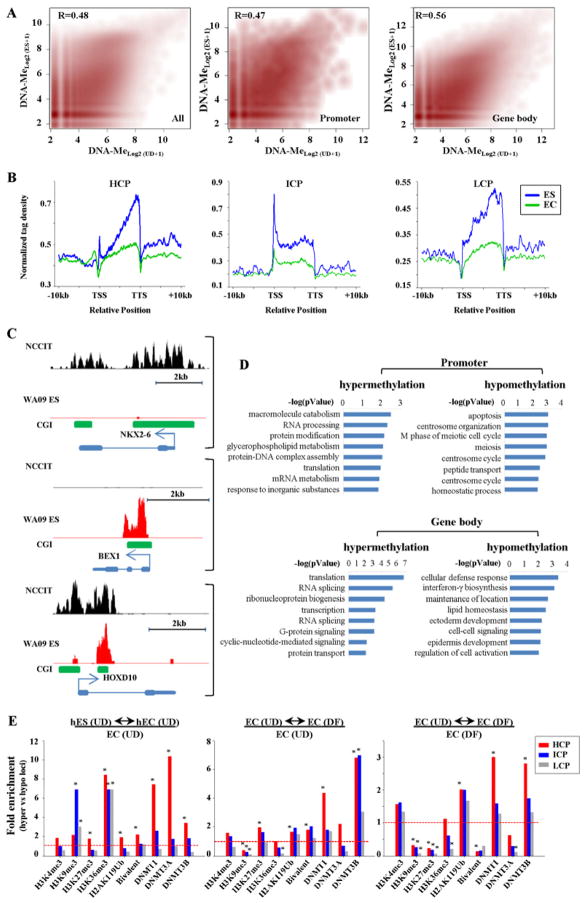
Figure 3. DNA methylation and DNMT localization in normal versus malignant stem cells and features of loci targeted for methylation.
(A) MBD-seq data from pluripotent NCCIT and WA09 human ES cells compared over the entire genome (left), promoters (middle), and gene bodies (right). Pearson correlation coefficients are shown. (B) Normalized tag distribution profiles of DNA methylation across gene bodies in undifferentiated hES and NCCIT cells. (C) Representative MBD-seq results in UD NCCIT (black) compared to WA09 human ES cells (red) at loci hypomethylated (top, NKX2-6), hypermethylated (middle, BEX1), or similarly methylated (bottom, HOXD10) in WA09 relative to NCCIT. (D) Biological processes enriched in differentially methylated genes in NCCIT (UD) compared to WA09 using DAVID. (E) Left panel. Enrichment of DNMTs, histone modifications, and bivalent status at loci hypermethylated in EC (undifferentiated) relative to ES, compared to those loci that are hypomethylated in EC relative to ES cells. Middle-right panels. Enrichment of DNMTs and histone marks at loci that become hypermethylated upon RA-induced differentiation of NCCIT cells, relative to those loci that become hypomethylated upon differentiation (4X or greater change). Enrichments are relative to the histone modifications and DNMT binding patterns derived from NCCIT in the undifferentiated condition in the middle panel and the differentiated condition in the right panel. Significance was assessed using the chi-square test (*p<0.01). Dashed line – 1X enrichment level.