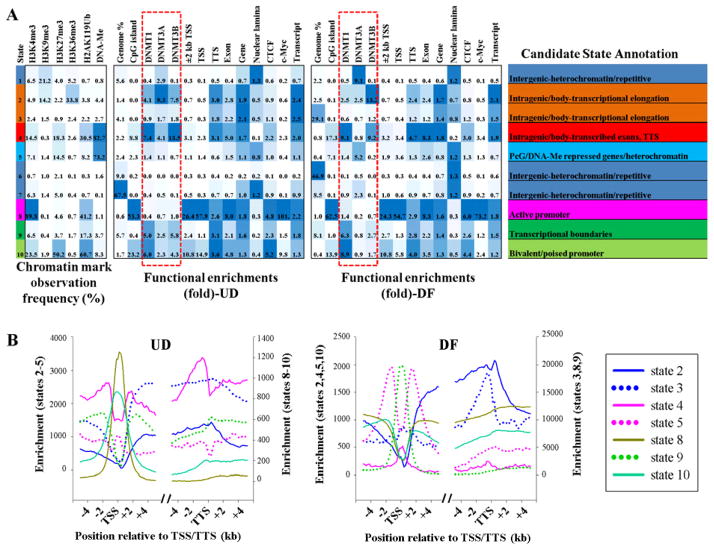
Figure 6. Discovery and characterization of chromatin states in NCCIT cells.
(A) A multivariate hidden Markov model was used to learn chromatin states jointly across undifferentiated and differentiated NCCIT cells. In the left part of the table, emission parameters that were learned de novo from recurrent combinations of epigenetic marks over the entire genome are shown. Each value represents the frequency of the epigenetic mark at genomic positions corresponding to that particular state. Functional enrichment of different features and candidate state annotations for each state in UD and DF conditions are shown on the right. Blue shading – intensity scaled by column. (B) Comparison of enrichments for states associated with genes (states 2–5, 8–10 only) across the TSS and the TTS regions in UD and DF cells. The break indicates the remainder of the gene body. States are graphed with different scales (right and left y-axes) to highlight their patterns, relative to other states, rather than absolute enrichments.