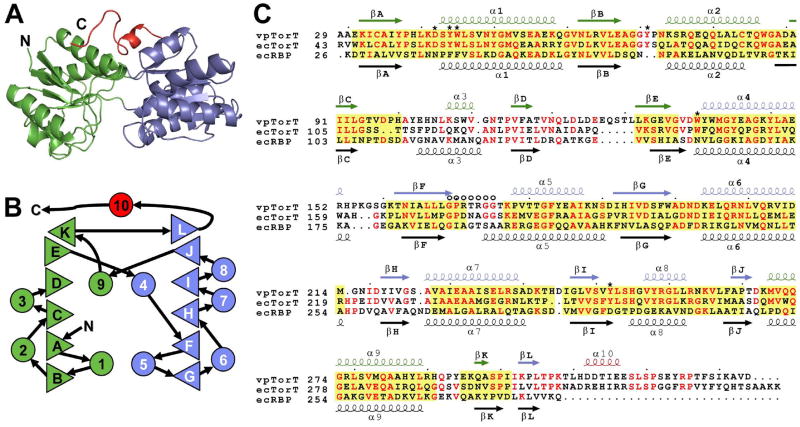
Figure 2. Structure-based TorT Sequence Alignments.
(A) Ribbon diagram of the vpTorT" structure, which uniquely includes a fully ordered βF-α5 loop. Coloring is by domain: N-terminal domain, green; C-terminal domain, blue; C-terminal extension, red.
(B) Topology diagram of vpTorT. Triangles represent β strands and circles represent α helices. Rightward- and leftward-facing triangles are for strands going into and coming out from the page, respectively. Color coding of these elements is as in A.
(C) Structure-based sequence alignment. Amino acid sequences of V. parahaemalyticus TorT (vpTorT), E. coli TorT (ecTorT), and E. coli ribose binding protein (ecRBP) are aligned based on structural superpositions of vpTorT and ecRBP structures and numbered accordingly. Secondary structure elements are symbolized and labeled above and below the respective sequences. Conserved residues are shown in red and structurally aligned regions where at least three contiguous Cα positions of ecRBP are within 3.0Å of vpTorTT, domain by domain, are shaded yellow. Residues that contact TMAO in TorTT and the βF-α5 loop are indicated by * and O, respectively.
See also Figure S3.