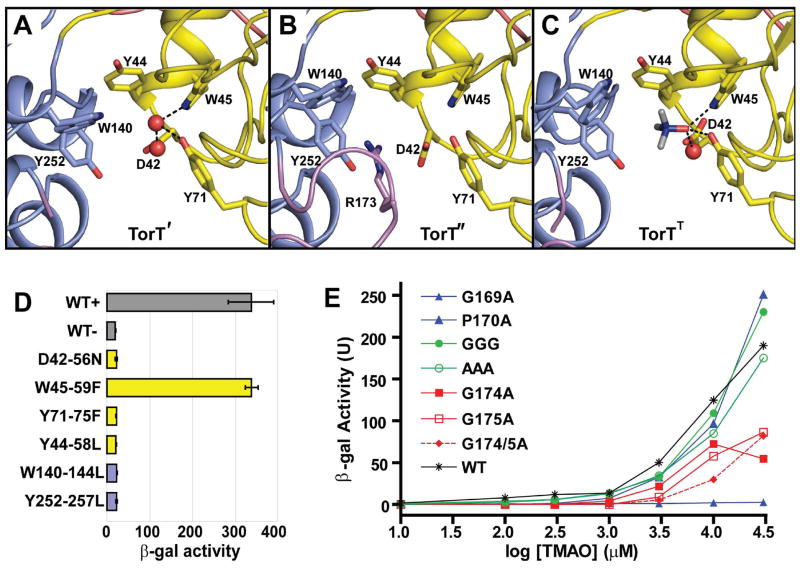
Figure 3. TMAO Binding Site and Mutational Analysis.
(A-C) Ribbon diagrams of the TMAO binding site TorT', TorT" and TorTT, respectively. The side chains of Asp42, Tyr44, Trp45, Tyr71, Trp140 and Tyr252 are identified in each. Arg173 is also labeled in TorT". Water molecules in the binding pockets of TorT' and TorTT are shown as red spheres. The NTD and CTD are colored yellow and blue, respectively, and ordered portions of the βF-α5 loops are colored magenta.
(D) β-Galactosidase activity of binding pocket mutants. Each culture was grown in the presence of 10 mM TMAO, except for WT-, which was grown in the absence of TMAO. Each mutant is labeled such that the vpTorT residue is first followed by the corresponding residue number in E. coli and the resulting mutation. The wild type (WT) results are shown in gray; mutations of NTD and CTD residues are in yellow and blue, respectively.
(E) β-Galactosidase activity of βF-α5 mutants at a series of TMAO concentrations. Wild type is in black (* T-S), mutations of the N-terminal glycine or proline residues are in blue, mutations of the three intervening residues into glycine (GGG) or alanine (AAA) are in green, and mutations of the C-terminal glycine residues are in red. Cited residue numbers are those from vpTorT whereas those for this segment in ecTorT are 5 greater, e.g. vpP170 becomes ecP175.
See also Table S2.