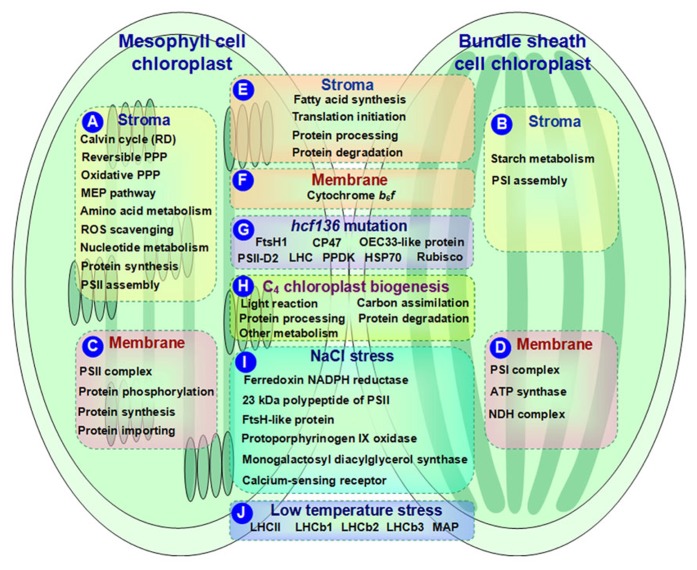
FIGURE 1.
Schematic presentation of cell-specific or stress-responsive pathways and proteins in maize chloroplasts revealed from proteomics studies. (A,B) Preferential metabolic pathways in the stroma of M and BS, respectively; (C,D) Preferential metabolic pathways and protein complexes in the chloroplast membrane of M and BS, respectively; (E,F) Metabolic pathways and protein complexes equally distributed in the chloroplast stroma/membrane of M and BS; (G) Differentially expressed proteins in chloroplasts of maize hcf136 mutant in comparison to wild-type; (H) Dynamics of metabolic pathways during maize chloroplast biogenesis; (I) Salt-responsive proteins in maize chloroplasts; (J) Low temperature-responsive proteins in maize chloroplasts. ATP, adenosine triphosphate; HSP, heat shock protein; LHC, light harvesting complex; MAP, minor antenna proteins; MEP, methylerythritol phosphate; NADPH, reduced nicotinamide adenine dinucleotide phosphate; NDH, NAD(P)H dehydrogenase; OEC, oxygen evolving center; PPDK, pyruvate orthophosphate dikinase; PPP, pentose phosphate pathway; PSI, photosystem I; PSII, photosystem II; RD, reductive phase; ROS, reactive oxygen species; Rubisco, ribulose-1,5-bisphosphate carboxylase/oxygenase.