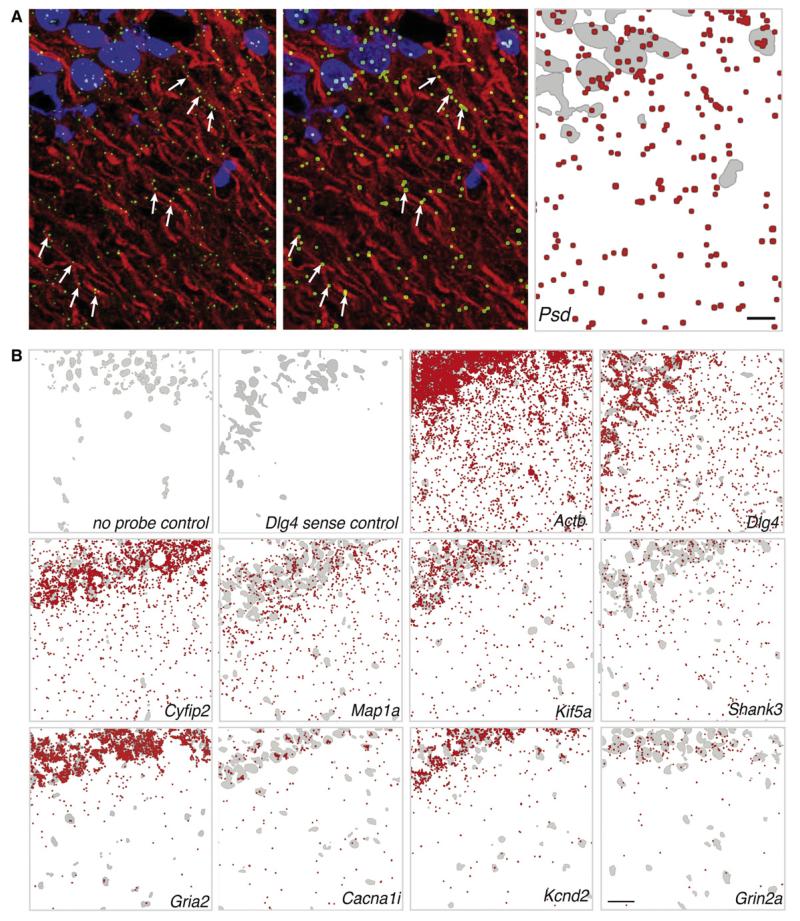
Figure 7. High-Resolution Fluorescent In Situ Hybridization Detection of mRNA Transcripts in the Synaptic Neuropil of Area CA1 of the Rat Hippocampus.
(A) Raw image from an in situ hybridization experiment using antisense probes against the Psd (pleckstrin and Sec7 domain protein) mRNA (green) in area CA1 of rat hippocampal slices (left). Dendrites were immunostained using an anti-Map2 antibody (red). The nuclei were stained with DAPI (blue). There are several examples where mRNA puncta are clearly present within the dendrites (arrows). In order to increase visibility when displaying larger areas of area CA1 we processed the images by dilating the mRNA puncta in ImageJ after setting a threshold (middle). For printing purposes, images were converted for display on white background (right). The DAPI channel was converted to a binary image, inverted, and displayed in gray to provide orientation in the CA1 region, the mRNA signal was converted to red, and the two channels were merged using Adobe Photoshop. Scale bar = 4 μm.
(B) Distribution of mRNA puncta in CA1 from in situ hybridization experiments in hippocampal slices displayed as indicated in (A) (right panel). In some control experiments, slices were not exposed to the probe (no probe) but to all subsequent amplification steps; in other controls, a sense probe followed by all subsequent amplification steps was used. Experiments were performed with the indicated probes. Scale bar = 25 μm.