Abstract
Bile acids (BAs) have many physiological roles and exhibit both toxic and protective influences within the liver. Alterations in the BA profile may be the result of disease induced liver injury. Nonalcoholic fatty liver disease (NAFLD) is a prevalent form of chronic liver disease characterized by the pathophysiological progression from simple steatosis to nonalcoholic steatohepatitis (NASH). The hypothesis of this study is that the ‘classical’ (neutral) and ‘alternative’ (acidic) BA synthesis pathways are altered together with hepatic BA composition during progression of human NAFLD. This study employed the use of transcriptomic and metabolomic assays to study the hepatic toxicologic BA profile in progressive human NAFLD. Individual human liver samples diagnosed as normal, steatosis, and NASH were utilized in the assays. The transcriptomic analysis of 70 BA genes revealed an enrichment of downregulated BA metabolism and transcription factor/receptor genes in livers diagnosed as NASH. Increased mRNA expression of BAAT and CYP7B1 were observed in contrast to decreased CYP8B1 expression in NASH samples. The BA metabolomic profile of NASH livers exhibited an increase in taurine together with elevated levels of conjugated BA species, taurocholic acid (TCA) and taurodeoxycholic acid (TDCA). Conversely, cholic acid (CA) and glycodeoxycholic acid (GDCA) were decreased in NASH liver. These findings reveal a potential shift toward the alternative pathway of BA synthesis during NASH, mediated by increased mRNA and protein expression of CYP7B1. Overall, the transcriptomic changes of BA synthesis pathway enzymes together with altered hepatic BA composition signify an attempt by the liver to reduce hepatotoxicity during disease progression to NASH.
Keywords: bile acids, liver, metabolomics, transcriptomics, nonalcoholic fatty liver disease, nonalcoholic steatohepatitis
Introduction
Hepatotoxicity in chronic liver disease is a result of alterations to hepatobiliary function, bile acid (BA) composition, and BA biosynthesis (Crosignani et al., 2007; Trottier et al., 2011; Trauner et al., 2008; Zollner et al., 2007). BAs are endogenous molecules that normally regulate cholesterol homeostasis, lipid solubilization and metabolic signaling (Li et al., 2011b; Rezen, 2011; Trauner et al., 2010). Exposure to increased levels of BAs in the diseased liver increases the risk of hepatotoxicity from activation of inflammatory, oxidative stress and necrotic cell death pathways (Tan et al., 2007; Allen et al., 2011). Nonalcoholic fatty liver disease (NAFLD) has become the most prevalent form of liver disease and is estimated to afflict 30-40% of the United States population (Ali et al., 2009). NAFLD is closely associated with features of the metabolic syndrome such as central girth obesity, insulin resistance and dyslipidemia (Dowman et al., 2010). This disease is particularly concerning to clinicians due to the potential for advancement in some patients to nonalcoholic steatohepatitis (NASH), which is characterized by extensive liver injury due to inflammation and fibrosis (McCullough, 2006). NASH is estimated to afflict up to 5.7-17% of the United States population and places patients at risk of further progression to cryptogenic cirrhosis and hepatocellular carcinoma (Dowman et al., 2010; McCullough, 2006; Sanyal et al., 2011). The mechanisms of disease progression in NAFLD are not fully understood and the contribution of altered BA composition and synthesis to the pathophysiological progression of the disease has not yet been characterized. In the current study we analyze altered BA synthesis in parallel with BA composition using transcriptomic and metabolomic methods in hepatic tissue samples representing the complete spectrum of NAFLD.
Changes in BA composition and synthesis can potentiate hepatotoxicity through pro-inflammatory mechanisms, membrane damage or cytotoxicity (Hofmann, 1999; Zhang et al., 2012; Zollner et al., 2007; Allen et al., 2011). BA synthesis occurs in the liver through the ‘classical’ (neutral) pathway and utilizes approximately 500 mg of cholesterol daily. Cholesterol is converted to primary BAs by several enzymes including CYP7A1, CYP8B1 and CYP27A1 (Thomas et al., 2008; Russell, 2003). A secondary BA synthesis pathway, the ‘alternative’ or oxysterol (acidic) pathway is also present and is reported to become more predominant and compensates for limitations of the classical pathway during disease (Crosignani et al., 2007). The alternative pathway utilizes extrahepatic cholesterol sources with the help of CYP27A1, CYP7B1 and CYP39A1 (Li et al., 2011b; Thomas et al., 2008). The primary BAs, cholic acid (CA) and chenodeoxycholic acid (CDCA), in addition to downstream secondary BAs generated by intestinal microflora make up the majority of the BA pool in humans (Li et al., 2011b). Synthesis contributes a limited amount of fresh BAs to the pool under normal conditions, because most BAs (95%) are efficiently conserved by enterohepatic recycling (Hofmann, 1999; Russell, 2003). Disease and other changes that affect the liver such as pregnancy, have been shown in animal models to alter enterohepatic recycling and BA synthesis through altered expresson of BA transporters and metabolizing enzymes (Aleksunes et al., 2012; Csanaky et al., 2009; Crosignani et al., 2007).
BA conjugation processes result in less toxic and more water-soluble BA species that are capable of protecting against damage from more toxic, hydrophobic BAs which can cause increased oxidative stress and cell death signaling (Perez et al., 2009; Delzenne et al., 1992; Thomas et al., 2008). It is estimated that 98% of hepatic BAs are conjugated to either taurine or glycine by the enzymes bile acid CoA synthase (BACS) and bile acid coenzyme A:amino acid N-acyltransferase enzyme (BAAT) (Russell, 2003). Liver defense mechanisms may also be activated through modifications in hepatic bile acid composition. Prior studies have analyzed the in vivo toxicological profiles of fed and endogenous BAs in rodent plasma and hepatic tissue and found increased conjugation (Zhang et al., 2012; Song et al., 2011). Altered plasma profiles of BA composition are also reported for many human chronic liver diseases including cholestasis, primary biliary cirrhosis, and NAFLD (Trottier et al., 2012; Kalhan et al., 2011). The purpose of this study is to determine whether hepatic BA composition and BA synthesis pathways are altered in the progression to human NASH as an adaptive attempt to reduce hepatocellular stress and toxicity. We have employed high-throughput metabolomic profiling techniques to characterize the altered BA profile and coupled that with a transcriptomic analysis of the classical and alternative (acidic) pathways of BA synthesis in NAFLD.
Materials and Methods
Human Liver Samples
Individual postmortem or biopsied human liver tissue samples were previously acquired from the Liver Tissue Cell Distribution System (LTCDS) funded by the National Institutes of Health. The LTCDS is comprised of the following institutions: the University of Minnesota, Virginia Commonwealth University and the University of Pittsburgh. Clinical information on these same liver tissue samples has been previously described in detail and published (Fisher et al., 2009). The classification of each liver sample was performed by an LTCDS medical pathologist using the NAFLD activity scoring (NAS)system (Kleiner et al., 2005). The samples were diagnosed by the scoring criteria as normal, steatosis, NASH (Fatty) or NASH (Not Fatty). The stage of steatosis was diagnosed as having >10% fat deposition within hepatocytes and no accompanying inflammation or fibrosis. NASH with fatty liver was characterized by >5% fat deposition in the presence of inflammation and fibrosis. NASH without fatty liver was distinguished by <5% fat deposition and increased inflammation and fibrosis. Normal (n=19), steatosis (n=10), NASH with fatty liver (n=9) and NASH without fatty liver (n=7) samples were utilized for mRNA isolation and application to Affymetrix 1.0 ST GeneChip microarrays as described previously (Lake et al., 2011). Samples reserved for metabolomic analysis included fewer normal (n=17) and steatosis (n=4) samples. An increased sample size of NASH with fatty liver (n=14) and NASH without fatty liver samples (n=23) were utilized in the metabolomic assays.
Human NAFLD Microarrays
Individual Affymetrix GeneChip Human 1.0 ST Arrays (Affymetrix, La Jolla, CA) were generated from purified mRNA for each NAFLD liver sample as described previously (Lake et al., 2011). A total of 33,252 genes for four pathologically defined groups (normal, steatosis, NASH fatty and NASH not fatty) are available in this array data set. It is publicly accessible through the ArrayExpress public repository for microarray data under the accession number E-MEXP-3291 (http://www.webcitation.org/5zyojNu7T). NASH fatty and NASH not fatty tissue categories were combined into one category for NASH due to the lack of mechanistic gene expression changes between these two pathologically differentiated NASH groups as explained previously for transcriptomic analyses (Lake et al., 2011).
Gene Set Enrichment Analysis
A total of 70 genes specifically implicated in hepatic BA synthesis were selected using literature database searches and examination of the Kyoto Encyclopedia of Genes and Genomes (KEGG) Database (http://www.kegg.jp/). Gene sets for BA metabolism, transport, and transcription factor/receptor were tested for enrichment among differentially expressed genes during NASH. All enrichment analyses were performed between two groups, a normal/steatosis combined group and the NASH group utilizing the generally applicable gene set enrichment analysis (GAGE) method (Luo et al., 2009).
Analytical Methods for LC/MS
A total of 17 healthy, 4 steatosis, 14 NASH (Fatty) and 23 NASH (Not Fatty) liver samples were weighed (weights recorded between 60 and 140 mg) and homogenized in 10 times the tissue weight of ice-cold methanol solution with 0.1% formic for 18-20 seconds using a polytron homogenizer over ice. Liver samples were kept frozen and on dry ice during all steps of the process. Samples were spun for 10 minutes at 4 degrees Celsius at 10000 RPM in a Beckman Coulter Allegra 25 centrifuge with a TA 10.250 rotor (Beckman Coulter Inc. Indianapolis, IN). Supernatants were transferred to new tubes, gently vortexed, and 200 microliter aliquots of each sample were added to the corresponding positions in a 96 deep well polypropylene plate (BrandTech Scientific, Inc. Essex, CT). The 96 well plate was dried using a V&P Scientific Model VP 177 96 well plate manifold dryer (V&P Scientific Inc. San Diego, CA) with nitrogen gas for approximately 6 hours prior to storage in a −80 degrees Celsius freezer and processing for UHPLC high resolution LC-MS analysis. Samples were reconstituted in a 90:10 water:methanol solution. D5-hippurate was added to each sample as an internal standard. The samples were then injected in randomized fashion onto a Thermo UHPLC Accela coupled to a Thermo Exactive high resolution orbitrap mass spectrometer. A total of 11 BA metabolites were acquired in negative ion mode and the organic sulfonic acid taurine was measured and acquired in positive ion mode (separate injections) with a mass accuracy within 5 ppm at 25K resolution. Metabolite peak areas under the curve (AUC) measurements for LC-MS were calculated using Component Elucidator, a software package developed by BMS scientists (Bristol-Myers Squibb Co, Princeton, NJ). The separation of DCA and CDCA using the conditions in this protocol was not possible and for the remainder of the analysis these BAs will be presented simultaneously. This study utilizes relative quantification of metabolites, a technique that is utilized in large, non-targeted profiling studies and normalizes metabolite intensities to that of an internal standard such as d5-hippurate.
Statistical Analysis of Metabolomics Data
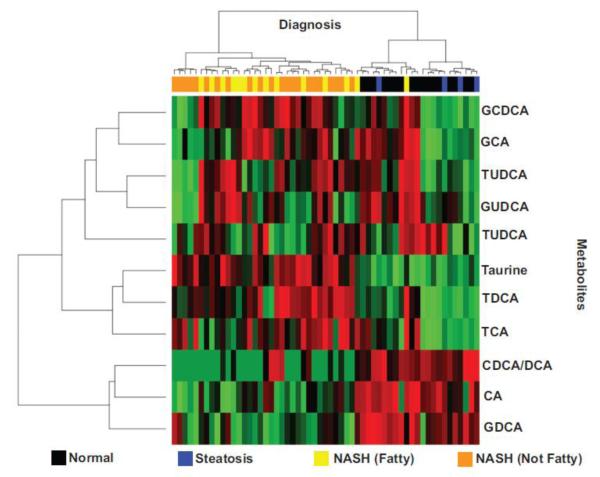
A total of 11 metabolites including the organic acid taurine were analyzed by the University of Arizona Statistical and Bioinformatics Consortium. The peak area under the curve (AUC) values for the LC-MS data were log transformed based upon a normal distribution approximation. After log transformation a one-way ANOVA with post hoc Tukey honest significant differences (HSD) testing was used for multiple comparisons of metabolites among the different diagnosis groups to test for mean differences. NASH (Fatty) and NASH (Not Fatty) diagnosis groups did not exhibit significant differences from one another in the analysis of BA metabolite levels. Thus, results for the two pathological categories were combined and presented as one group designated as NASH. Significance was defined as p values ≤ 0.05 and represented by an asterisk (*) if significantly changed from normal samples and a pound sign (#) if significantly changed from steatosis samples (Figure 3). Percent of normal values were calculated using the raw metabolite data of the steatosis and NASH sample groups. Hierarchical clustering analysis was performed on the metabolite data to reveal clustering effects related to the stage of the disease.
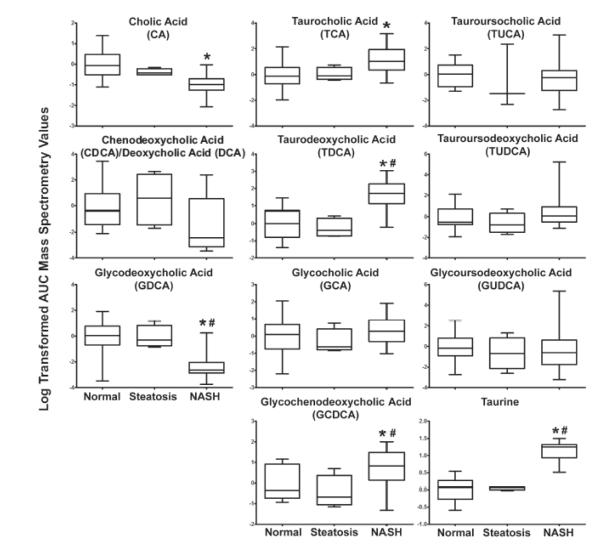
Figure 3. Bile Acid Metabolite Levels in Hepatic Samples of Human NAFLD.
The peak area under the curve (AUC) measurement for each metabolite is revealed as log transformed mass spectrometry values +/− the standard deviation. Significance is defined as p values ≤ 0.05. A one way ANOVA with a Tukey post hoc test of mean normalized metabolites was utilized to determine statistical significance. Asterisk (*) represents significance from normal, pound sign (#) represents significance from steatosis. Sample size: normal n=17, steatosis n=5, NASH (Fatty) n=14, NASH (Not Fatty) n=23.
Immunoblot Protein Analysis
Western blots were performed to validate the protein expression of CYP7B1, CYP8B1 and BAAT in normal (n=7), steatosis (n=7), and NASH Fatty (n=8) and NASH Not Fatty (n=7) whole cell liver lysate samples. Samples were prepared at a concentration of 80 micrograms in Laemmli sample dye (Bio-Rad Laboratories, Hercules, CA) with β-mercaptoethanol and boiled for 10 minutes. Proteins were separated on 10% SDS polyacrylamide gels and transferred to methanol activated polyvinylidene difluoride (PVDF) membranes (Bio-Rad, Hercules, CA) at 30 milliamps for 12.5 hours. Primary antibodies were acquired from Abcam Inc. (Cambridge, MA) and Santa Cruz Biotechnology (Santa Cruz, CA). CYP8B1 (sc-101387) was used in 5% nonfat dry milk at a concentration of 1:7,000 with 1:100,000 secondary antibody (sc-2005). BAAT (ab97455) and CYP7B1 (ab77157) antibodies were utilized at primary concentrations of 1:7,000 in TBST with secondary antibody concentrations of 1:100,000 (sc-2004) and 1:200,000 (sc-2302) respectively. For total protein control pan-Cadherin primary antibody was used at a concentration of 1:7,000 (ab16505) (Abcam, Cambridge, MA). Blots were imaged with advanced chemiluminescence substrate (GE Healthcare, Piscataway, NJ). Relative protein expression was determined using densitometric Image J software (National Institutes of Health, Bethesda, MD). All proteins were normalized to total pan-cadherin protein. The pathological stages of NASH (Fatty) and NASH (Not Fatty) did not exhibit significant changes from each other, and results are presented as one combined group designated as NASH (n=15). Statistical significance was determined by one way ANOVA with a Tukey post hoc testing analysis in GraphPad Prism 5 software (La Jolla, CA).
Spearman Correlation Analysis of BA Metabolites and Genes
The correlations between the levels of BA metabolites and BA gene expression were calculated by Spearman tests. Corresponding p values were computed using the algorithm AS 89 (Best et al., 1975). P value corrections for the multiple testing were adjusted according to the methods of Benjamini and Hochberg.
Results
Bile Acid Gene Set Enrichment Analysis
Genes divided into BA metabolism, transporter and transcription factor/receptor categories were analyzed for gene set enrichment using the previously published human NAFLD microarray data set (Lake et al., 2011). The purpose of the enrichment testing was to demonstrate which gene categories were more frequently up or downregulated when compared against a background set of genes (Table 1). The gene set enrichment analysis revealed that the BA gene category containing all 70 genes was on average enriched for downregulated genes during NASH (Table 1). BA metabolism and transcription factor/receptor categories were enriched for downregulation in samples diagnosed as NASH. Conversely, the number of differentially expressed genes within the BA transport gene category did not exhibit significant deviation from the array average (Table 1 and Supplemental Table 1).
Table 1. Gene Enrichment Analysis.
Gene set enrichment testing of BA gene categories for either up or downregulated gene expression is shown. A statistically enriched gene set is defined as p values ≤ 0.05. The false discovery rate (FDR) q value is the Benjamini and Hochberg adjusted p value.
| Downregulation Enrichment Analysis | |||
|---|---|---|---|
| Gene Category | P Value | FDR q Value | Gene Set Size |
| Bile Acid Transport | 0.260 | 0.260 | 34 |
| Bile Acid Metabolism | 0.040 | 0.054 | 26 |
| Bile Acid Receptors and Transcription Factors |
0.012 | 0.029 | 21 |
| All Bile Acid Genes | 0.015 | 0.029 | 81 |
| Upregulation Enrichment Analysis | |||
| Bile Acid Transport | 0.740 | 0.988 | 34 |
| Bile Acid Metabolism | 0.960 | 0.988 | 26 |
| Bile Acid Receptors and Transcription Factors |
0.988 | 0.988 | 21 |
| All Bile Acid Genes | 0.985 | 0.988 | 81 |
Gene Expression of Individual Bile Acid Metabolizing Enzymes
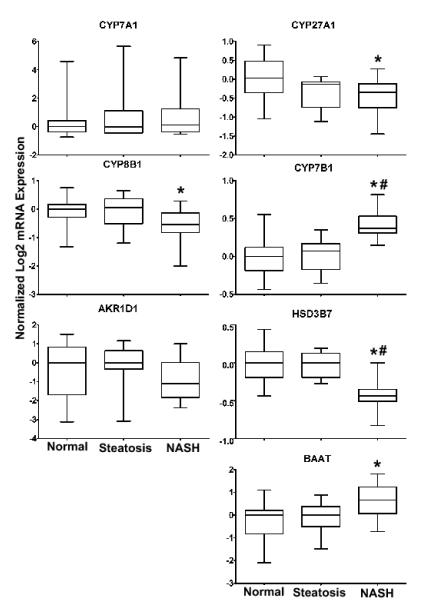
The mRNA expression levels for the enzymes CYP7A1, CYP27A1, CYP7B1, CYP8B1, AKR1D1, HSD3B7 and BAAT were analyzed using the log transformed human microarray data (Figure 1). CYP27A1 and HSD3B7 genes were significantly decreased in NASH liver samples while CYP7A1 and AKR1D1 did not change. CYP7B1 mRNA levels were significantly increased in expression during NASH when compared to normal or steatosis (Figure 1). This upregulation of CYP7B1 is consistent with the regulation of the alternative (acidic) BA synthesis pathway during NASH (Figure 1 and 6). The conjugating enzyme BAAT also exhibited a significant increase, while in contrast CYP8B1 was significantly decreased in NASH liver samples.
Figure 1. Bile Acid mRNA Expression.
The mRNA expression of classical and alternative BA synthesis pathways enzymes were analyzed from the human NAFLD microarrays. Data are presented as the log2 mRNA expression +/− the standard deviation. A one way ANOVA with a Tukey post hoc test of mean normalized log2 mRNA expression was utilized to determine statistical significance. Significance is defined as p values ≤ 0.05. An asterisk (*) represents significance from normal and a pound sign (#) represents significance from steatosis.
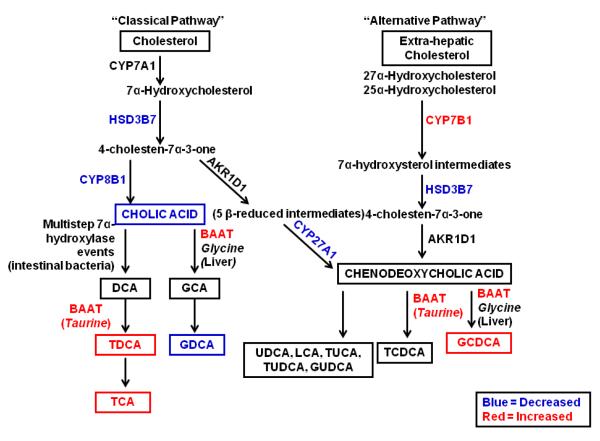
Figure 6. Bile Acid Synthesis in Human NASH.
A simplified schematic of the hepatic classical and alternative pathways of BA synthesis in human NASH is provided. Increased levels of metabolites (TCA, TDCA, GCDCA and taurine) are shown in red. Gene expression increases of metabolizing enzymes (BAAT, CYP7B1) are also shown in red. Decreased metabolite levels (CA, GDCA) and enzyme gene expression (CYP27A1, CYP8B1, and HSD3B7) is represented in blue. Unchanged metabolites (CDCA/DCA, GUDCA, TUCA, TUDCA, GCA) and enzymes (CYP7A1, AKR1D1) are shown in black. Note: Not every enzymatic step in BA synthesis and systemic enterohepatic recycling of BAs is shown in this schematic. This pathway is adapted and modified from (Beilke et al., 2009) and (Thomas et al., 2008).
Hierarchical Clustering Analysis
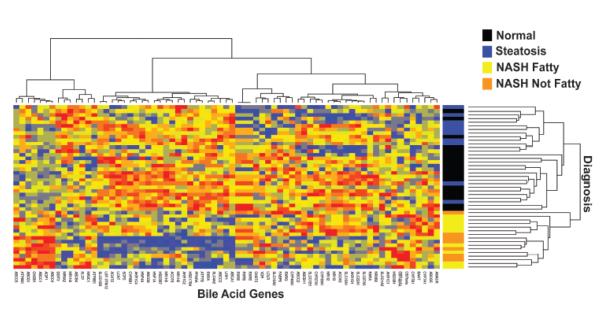
An unsupervised hierarchical clustering analysis demonstrated that expression changes of the BA genes were sufficient to distinguish NASH from normal and steatosis diagnosis groups (Figure 2). Clustering revealed two major groups in the diagnosis groups on the heatmap. One cluster corresponded to NASH samples while the other corresponded to a mixture of normal and steatosis samples (Figure 2).
Figure 2. Hierarchical Clustering Analysis of Bile Acid Genes in Human NAFLD.
The heatmap demonstrates the changes in expression for all BA genes in normal, steatosis and NASH (Fatty) and NASH (Not Fatty) samples. Both genes and samples were sorted using unsupervised hierarchical clustering. Red and blue colors correspond to increased and decreased genes respectively. Yellow color correlates to the median expression. Clustering of diagnosis groups with respect to gene expression changes is shown.
Bile Acid Metabolomics
The metabolomics analysis of hepatic BAs measured by LC-MS revealed significant changes among samples diagnosed as NASH (Figure 3, Supplemental Tables 2 & 3). The average area under the curve (AUC) measurements for cholic acid (CA) exhibited a significant decrease in liver samples diagnosed as NASH (31% of normal) while glycodeoxycholic acid (GDCA) was significantly decreased to 9% of normal in NASH (Figure 3, Supplemental Tables 2 & 3). Conversely, significant increases in the metabolite levels of taurocholic acid (TCA) (298% of normal), taurodeoxycholic acid (TDCA) (507% of normal), and glycochenodeoxycholic acid (GCDCA) (197% of normal) were observed in samples diagnosed as NASH. Glycoursodeoxycholic acid (GUDCA), tauroursocholic acid (TUCA) and tauroursodeoxycholic acid (TUDCA) did not show any statistically significant differences between diagnosis categories. The organic sulfonic acid, taurine, showed a significant increase to 304% of normal in NASH samples (Figure 3, Supplemental Tables 2 & 3). However, no statistically significant changes were observed in steatosis when compared to normal samples for taurine (Figure 3, Supplemental Table 3). A hierarchical clustering analysis of rank-transformed AUC values revealed that metabolite levels of these bile acids and taurine distinguish NASH samples from those diagnosed as normal or steatosis. Samples diagnosed as NASH formed a cluster together according to BA composition changes (Figure 4). The biochemical observations of the decreased classical pathway BAs (CA and GDCA) and increased taurine-conjugated BA species (TDCA and TCA) are consistent with the transcriptional findings (Figure 1 and 6).
Figure 4. Hierarchical Clustering Analysis of Metabolites.
The heatmap of BA metabolites demonstrates the increase or decrease of each metabolite in liver samples diagnosed as either normal, steatosis, NASH (Fatty) and NASH (Not Fatty). Rank transformed metabolite levels and samples were sorted using unsupervised hierarchical clustering. High, medium and low levels of metabolites are shown in red, black and green colors respectively.
Correlation Analysis of Bile Acid Genes and Metabolites
Correlation testing for associations between metabolite and mRNA levels was performed to reveal the relationship between transcriptomic and metabolomic data sets. Metabolites and mRNA levels were both rank-transformed to perform a Spearman correlation analysis in order to determine whether expression levels of mRNA correlated with bile acid metabolite levels. To correct for multiple testing, p values were adjusted according to Benjamini and Hochberg (Table 2). CA, DCA and GDCA revealed significantly negative correlations [−0.5843 (p value=0.0057), −0.5489 (p value=0.012), −0.6013 (p value=0.004)] with CYP7B1 gene expression by Spearman correlation testing (Table 2). Significant positive correlations between CYP7B1 and taurine [0.6838 (p value=0.000581)] as well as CYP8B1 and GDCA [0.4733 (p value=0.0412)] were observed in the correlation tests for normal to NASH samples (Table2). Correlations of all 70 BA genes with metabolites are shown in supplemental data (Supplemental Table 2).
Table 2. Spearman Rank Correlations and Adjusted P Values of Selected Bile Acid Genes and Metabolites.
The Spearman rank correlation values of three critical BA pathway genes and metabolites with the corresponding adjusted p values (in parentheses) is shown. Statistical significance of the correlation is represented by p value ≤ 0.05 and is designated by asterisk (*).
| Metabolites | CYP7B1 | BAAT | CYP8B1 |
|---|---|---|---|
| TCA | 0.2664 (0.343) | 0.2661 (0.343) | 0.0664 (0.858) |
| TDCA | 0.4476 (0.0624) | 0.2507 (0.383) | −0.2048 (0.475) |
| CA | −0.5843 (0.0057)* | −0.3572 (0.164) | 0.3062 (0.259) |
| DCA/CDCA | −0.5489 (0.012)* | −0.4068 (0.0936) | 0.1638 (0.581) |
| GDCA | −0.6013 (0.004)* | −0.4478 (0.0601) | 0.4733 (0.0412)* |
| GCA | 0.1445 (0.636) | 0.3369 (0.199) | 0.3347 (0.202) |
| GCDCA | 0.3524 (0.169) | 0.3159 (0.238) | −0.0301 (0.936) |
| TUCA | −0.0574 (0.864) | −0.2037 (0.475) | 0.2138 (0.458) |
| TUDCA | 0.2529 (0.38) | 0.0707 (0.851) | −0.0284 (0.942) |
| GUDCA | 0.0272 (0.943) | −0.0036 (0.994) | 0.2188 (0.68) |
| Taurine | 0.6838 (0.000581)* | 0.3936 (0.107) | −0.3741 (0.132) |
Protein Expression of Bile Acid Synthesis Enzymes
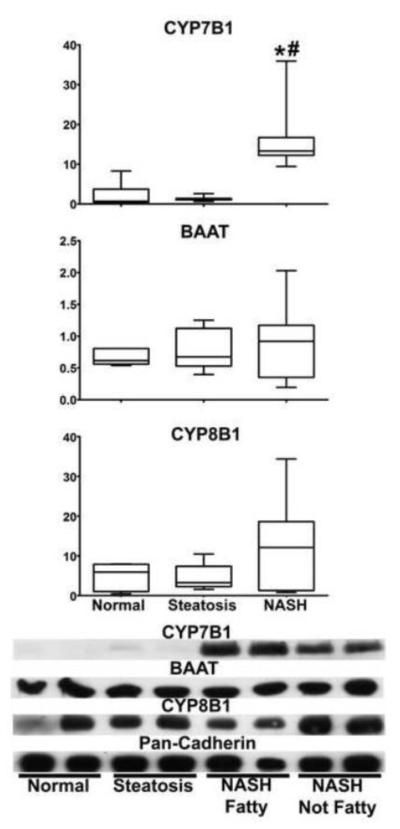
The protein expression levels of CYP7B1, CYP8B1 and BAAT were analyzed by western blot analysis in normal, steatosis and NASH human liver whole cell lysates with pan-cadherin total protein. CYP7B1 was significantly increased in NASH samples compared to normal and steatosis (Figure 5). The relative protein levels of CYP8B1 and BAAT were not significantly altered at any stage of NAFLD (Figure 5).
Figure 5. Protein Expression of Bile Acid Synthesis Enzymes.
The relative protein expression of three key bile acid synthesis enzymes (CYP7B1, BAAT and CYP8B1) in human whole cell liver lysate samples is shown. Representative blots with two samples per diagnosis group are given. The relative protein expression of all samples was analyzed by densitometric analysis and normalized to pan-cadherin total protein. A one way ANOVA with a Tukey post hoc test of mean normalized relative protein expression was utilized to determine statistical significance. Significance is defined as p values ≤ 0.05. Asterisk (*) represents significance from normal and pound sign (#) represents significance from steatosis.
Discussion
In the current study, we have demonstrated altered BA composition and a shift from the classical to the alternative BA synthesis pathway in human livers diagnosed as NASH (Figure 3 and 6). Alterations in BA composition and synthesis are important mechanisms that modulate hepatotoxicity. The accumulation of BAs in the liver and the associated toxicity is a significant problem in many diseases (Trottier et al., 2011; Trottier et al., 2012; Perez et al., 2009; Crosignani et al., 2007). Evidence of BA toxicity is abundant and results from excessive exposure to either primary or secondary BAs (Delzenne et al., 1992; Hofmann, 1999; Song et al., 2011). While the mechanisms of BA injury in NASH are not completely understood, it is established that oxidative stress and the antioxidant response play a role in the pathological progression to NASH (Hardwick et al., 2010). Hydrophobic BAs are known to impair the mitochondrial electron transport chain resulting in massive ROS production and oxidative stress. Increased exposure to hydrophobic BAs also results in the direct activation of apoptosis and necrosis pathways (Perez et al., 2009; Sharma et al., 2010). BAs are also reported to act as inflammatory mediators in rodent liver injury models (Zhang et al., 2012) and isolated hepatocyte experiments (Allen et al., 2011). Furthermore, BAs are capable of activating stellate cells and profibrogenic mechanisms (Svegliati-Baroni et al., 2005) which are both pathological components of NASH. The hydrophobicity, and thus the potential for BA toxicity has been reported in the following order: DCA>CDCA>CA>UDCA (Perez et al., 2009; Song et al., 2011). Hydrophobic BAs are recognized as the main precursors of many toxic events in the liver however, some BA species are actually beneficial. TDCA, and TUDCA have been shown in the literature to protect organs against oxidative and endoplasmic reticulum stress (Ratziu et al., 2011; Seyhun et al., 2011; Yang et al., 2010). In the present study we demonstrate a significant increase in metabolite levels of more hydrophilic BA components in NASH samples (TCA, TDCA, GCDCA and GCA). The concurrent decrease of CA and GDCA in NASH altogether is suggestive of an attempt in these livers to reduce the profile of more toxic hydrophobic BAs (Figure 3 and 6).
The changes in BA levels in NASH depend upon the functional activation or suppression of enzymes in the classical and alternative BA synthesis pathways. We discovered an enrichment of downregulated BA metabolism enzymes and transcription factor/receptors in NASH livers. Similarly, rodent studies examining the role of pregnancy in BA alterations have also found decreased mRNA levels of many hepatic transcription factors and some BA metabolizing enzymes (Aleksunes et al., 2012). The enrichment of downregulated BA metabolism enzymes in human NASH liver included the classical pathway enzyme, CYP8B1 that is similarly suppressed in farnesoid x receptor (fxr) activated mice (Kong et al., 2012). The transcriptional decrease of CYP8B1 in NASH liver may be indicative of an attempt to decrease production of CA through the classical pathway and has also been observed in animal models of cholestatic liver injury (Zhang et al., 2012). The increased mRNA and protein levels of CYP7B1 are indicative of a functional upregulation of the alternative (acidic) pathway in NASH liver (Figure 1 and 5). Furthermore, the transcriptional increase of BAAT in NASH suggests an attempt by the liver to increase BA conjugation in the liver. These findings in the BA gene expression data in human NASH are similar to results in a previous study that examined the BA metabolism pathway in a rodent model of cholestasis (Beilke et al., 2008). The lithocholic acid (LCA) induced cholestasis mouse model utilized in our laboratory also explored changes in metabolism pathway enzymes and BA composition (Beilke et al., 2009; Beilke et al., 2008). Mice treated with phenobarbital or 1,4-bis[2-(3,5-dichloropyridyloxy)] benzene (TCPOBOP) before LCA exposure exhibited induced expression of constitutive androstane receptor (CAR). The induction of CAR in these mice also resulted in subsequent alterations in BA composition. The gene expression of Cyp7b1 was significantly upregulated in these mice and parallels what is seen in human NASH. Additionally, increased mRNA expression of the conjugation enzyme Baat was also observed in the CAR-activated mice compared to mice lacking CAR expression. The gene expression levels of BA metabolism enzymes for both CYP8B1, CYP7B1 and BAAT in human NASH liver is supported by these previous studies in animal models of liver disease (Beilke et al., 2008; Beilke et al., 2009; Kong et al., 2012). The altered expression of BA metabolism genes aid in a shift from the classical pathway of BA synthesis to the alternative and is indicative of a defense mechanism altering the overall BA profile to protect against hepatotoxicity. This finding is similar to the observations of shifted BA synthesis and composition in the rodent model of LCA-induced cholestasis (Beilke et al., 2009).
Shifts in BA synthesis and composition have been reported in various human hepatic disease states such as primary biliary cirrhosis (PBC) and chronic hepatitis C (Crosignani et al., 2007; Crosignani et al., 2011). The classical pathway of BA synthesis is reported to be impaired in these patients. In contrast, alternative pathway intermediates are intact in these patients as represented by normal levels of 27α-hydroxycholesterol in plasma (Crosignani et al., 2011). Studies in obese patients have also shown an upregulation of the alternative pathway (Crosignani et al., 2011). The findings of these clinical studies support the results observed in this study of NASH.
In addition to the increased levels of taurine-conjugated BAs (TCA and TDCA) in NASH livers, a concurrent increase in the levels of the organic acid taurine were observed. Elevated levels of taurine in serum have been previously reported in studies of NASH patients (Li et al., 2011a). Additionally, recent pre-clinical and clinical studies support the use of taurine administration to alleviate inflammation and lipid accumulation in NAFLD (Timbrell et al., 1995; Chen et al., 2006; Gentile et al., 2011). Increased concentrations of taurine-conjugated BAs in cholestatic liver disease are reported in the literature and parallel our findings (Trottier et al., 2012). Additionally, elevated levels of GCDCA and TCA have been reported in the plasma of NASH patients, corresponding to the increases seen in our hepatic NASH samples (Kalhan et al., 2011). The elevated levels of taurine and conjugated BA species are indicative of an initiated protective mechanism by the liver to alter BA composition and compensate for disease induced hepatotoxicity during NASH. The beneficial aspects of taurine conjugated BAs are reported in the literature (Hirano et al., 2006; Seyhun et al., 2011) along with the exploration of taurine as a potential therapeutic in human NAFLD (Gentile et al., 2011). Composition patterns of BAs in human bile among different states of liver disease also demonstrate a coordinated pattern of change among glycine and taurine conjugated BAs (Nagana et al., 2009) which also support our findings of altered BA composition and increased levels of conjugated BAs in the NASH hepatic samples.
In summary, the transcriptomic results and metabolomic BA profile provide evidence for a transition from the classical to the alternative pathway of BA synthesis in the livers of NASH patients (Figure 6). The increased availability of hepatic taurine for bile acid conjugation reactions is suggestive of an attempt to ameliorate the potential toxic effects of endogenous BAs in NASH. These results clearly show altered BA composition in the form of increased levels of taurine-conjugated BAs and decreased levels of CA and GDCA. Increased protein and mRNA levels of CYP7B1 with the corresponding decrease in gene expression of CYP8B1 indicate the initiation of mechanisms at the functional and transcriptional level acting to shift the pathways in NASH. The unique analytical approach of combining transcriptomic and metabolomic findings demonstrates the importance of gene expression changes upon the biochemical outcome in these patients. In conclusion, the data presented here, provide important new insights into the molecular and biochemical responses to NAFLD progression and provide the foundation for further evaluation of the contribution of altered BA metabolism in the development of NASH.
Supplementary Material
Highlights.
Altered hepatic bile acid composition is observed in progressive NAFLD.
Bile acid synthesis enzymes are transcriptionally altered in NASH livers.
Increased levels of taurine and conjugated bile acids are observed in NASH.
Hepatic bile acid synthesis shifts toward the alternative pathway in NASH.
Acknowledgements
We extend our sincere gratitude to Dr. Walter T. Klimecki for his valued scientific advice and contribution to the development of the human NAFLD microarray data set. We also thank the NIH-sponsored Liver Tissue Cell Distribution System members for their help in the acquisition of human liver tissue at the University of Minnesota, Virginia Commonwealth and the University of Pittsburgh.
Funding This work was supported by the National Institutes of Health Grants [DK068039, AI083927, AT002842, HD062489, ES006694], the National Institute of Environmental Health Science Toxicology Training Grant [ES007091]. The Academy of Sciences of the Czech Republic [AVOZ50510513]. The Liver Tissue Cell Distribution System. National Institute of Health Contract [NO1-DK-7-0004/HHSN267200700004C].
Abbreviations
- NAFLD
Nonalcoholic fatty liver disease
- NASH
nonalcoholic steatohepatitis
- BA
bile acid
- CA
cholic acid
- DCA
deoxycholic acid
- CDCA
chenodeoxycholic acid
- TCA
taurocholic acid
- TDCA
taurodeoxycholic acid
- GCA
glycocholic acid
- GDCA
glycodeoxycholic acid
- GCDCA
glycochenodeoxycholic acid
- GUDCA
glycoursodeoxycholic acid
- TUDCA
tauroursodeoxycholic acid
- TUCA
tauroursocholic acid
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- 1.Aleksunes LM, Yeager RL, Wen X, Cui JY, Klaassen CD. Repression of Hepatobiliary Transporters and Differential Regulation of Classic and Alternative Bile Acid Pathways in Mice During Pregnancy. Toxicological Sciences. 2012;130:257–268. doi: 10.1093/toxsci/kfs248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ali R, Cusi K. New diagnostic and treatment approaches in non-alcoholic fatty liver disease (NAFLD) Annals of Medicine. 2009;41:265–278. doi: 10.1080/07853890802552437. [DOI] [PubMed] [Google Scholar]
- 3.Allen K, Jaeschke H, Copple BL. Bile Acids Induce Inflammatory Genes in Hepatocytes: A Novel Mechanism of Inflammation during Obstructive Cholestasis. The American Journal of Pathology. 2011;178:175–186. doi: 10.1016/j.ajpath.2010.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beilke LD, Aleksunes LM, Holland RD, Besselsen DG, Beger RD, Klaassen CD, Cherrington NJ. Constitutive androstane receptor-mediated changes in bile acid composition contributes to hepatoprotection from lithocholic acid-induced liver injury in mice. Drug Metabolism and Disposition. 2009;37:1035–1045. doi: 10.1124/dmd.108.023317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beilke LD, Besselsen DG, Cheng Q, Kulkarni S, Slitt AL, Cherrington NJ. Minimal role of hepatic transporters in the hepatoprotection against LCA-induced intrahepatic cholestasis. Toxicological Sciences. 2008;102:196–204. doi: 10.1093/toxsci/kfm287. [DOI] [PubMed] [Google Scholar]
- 6.Best DJ, Roberts DE. Algorithm AS91. Percentage points of the chi-squared distribution. Applied Statistics. 1975:385–388. [Google Scholar]
- 7.Chen SW, Chen YX, Shi J, Lin Y, Xie WF. The restorative effect of taurine on experimental nonalcoholic steatohepatitis. Digestive Diseases and Sciences. 2006;51:2225–2234. doi: 10.1007/s10620-006-9359-y. [DOI] [PubMed] [Google Scholar]
- 8.Crosignani A, Del Puppo M, Longo M, De Fabiani E, Caruso D, Zuin M, Podda M, Javitt NB, Kienle MG. Changes in classic and alternative pathways of bile acid synthesis in chronic liver disease. Clinica Chimica Acta. 2007;382:82–88. doi: 10.1016/j.cca.2007.03.025. [DOI] [PubMed] [Google Scholar]
- 9.Crosignani A, Zuin M, Allocca M, Del Puppo M. Oxysterols in bile acid metabolism. Clinica Chimica Acta. 2011;412:2037–2045. doi: 10.1016/j.cca.2011.07.028. [DOI] [PubMed] [Google Scholar]
- 10.Csanaky IL, Aleksunes LM, Tanaka Y, Klaassen CD. Role of hepatic transporters in prevention of bile acid toxicity after partial hepatectomy in mice. American Journal of Physiology - Gastrointestinal and Liver Physiology. 2009;297:G419–G433. doi: 10.1152/ajpgi.90728.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delzenne NM, Calderon PB, Taper HS, Roberfroid MB. Comparative hepatotoxicity of cholic acid, deoxycholic acid and lithocholic acid in the rat: in vivo and in vitro studies. Toxicology Letters. 1992;61:291–304. doi: 10.1016/0378-4274(92)90156-e. [DOI] [PubMed] [Google Scholar]
- 12.Dowman JK, Tomlinson JW, Newsome PN. Pathogenesis of non-alcoholic fatty liver disease. QJM: Monthly Journal of the Association of Physicians. 2010;103:71–83. doi: 10.1093/qjmed/hcp158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fisher CD, Lickteig AJ, Augustine LM, Ranger-Moore J, Jackson JP, Ferguson SS, Cherrington NJ. Hepatic Cytochrome P450 Enzyme Alterations in Humans with Progressive Stages of Nonalcoholic Fatty Liver Disease. Drug Metabolism and Disposition. 2009;37:2087–2094. doi: 10.1124/dmd.109.027466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gentile CL, Nivala AM, Gonzales JC, Pfaffenbach KT, Wang D, Wei Y, Jiang H, Orlicky DJ, Petersen DR, Pagliassotti MJ, Maclean KN. Experimental evidence for therapeutic potential of taurine in the treatment of nonalcoholic fatty liver disease. American Journal of Physiology - Regulatory, Integrative and Comparative Physiology. 2011;301:R1710–R1722. doi: 10.1152/ajpregu.00677.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hardwick RN, Fisher CD, Canet MJ, Lake AD, Cherrington NJ. Diversity in antioxidant response enzymes in progressive stages of human nonalcoholic fatty liver disease. Drug Metabolism and Disposition. 2010;38:2293–2301. doi: 10.1124/dmd.110.035006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hirano F, Haneda M, Makino I. Chenodeoxycholic acid and taurochenodexycholic acid induce anti-apoptotic cIAP-1 expression in human hepatocytes. Journal of Gastroenterology and Hepatology. 2006;21:1807–1813. doi: 10.1111/j.1440-1746.2006.04363.x. [DOI] [PubMed] [Google Scholar]
- 17.Hofmann AF. Bile acids: the good, the bad, and the ugly. Physiology. 1999;14:24–29. doi: 10.1152/physiologyonline.1999.14.1.24. [DOI] [PubMed] [Google Scholar]
- 18.Kalhan SC, Guo L, Edmison J, Dasarathy S, McCullough AJ, Hanson RW, Milburn M. Plasma metabolomic profile in nonalcoholic fatty liver disease. Metabolism. 2011;60:404–413. doi: 10.1016/j.metabol.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kleiner DE, Brunt EM, Van Natta M, Behling C, Contos MJ, Cummings OW, Ferrell LD, Liu YC, Torbenson MS, Unalp-Arida A, Yeh M, McCullough AJ, Sanyal AJ. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology. 2005;41:1313–1321. doi: 10.1002/hep.20701. [DOI] [PubMed] [Google Scholar]
- 20.Kong B, Wang L, Chiang JYL, Zhang Y, Klaassen CD, Guo GL. Mechanism of tissue-specific farnesoid X receptor in suppressing the expression of genes in bile-acid synthesis in mice. Hepatology. 2012;56:1034–1043. doi: 10.1002/hep.25740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lake AD, Novak P, Fisher CD, Jackson JP, Hardwick RN, Billheimer DD, Klimecki WT, Cherrington NJ. Analysis of global and absorption, distribution, metabolism, and elimination gene expression in the progressive stages of human nonalcoholic fatty liver disease. Drug Metabolism and Disposition. 2011;39:1954–1960. doi: 10.1124/dmd.111.040592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li H, Wang L, Yan X, Liu Q, Yu C, Wei H, Li Y, Zhang X, He F, Jiang Y. A proton nuclear magnetic resonance metabonomics approach for biomarker discovery in nonalcoholic fatty liver disease. Journal of Proteome Research. 2011a;10:2797–2806. doi: 10.1021/pr200047c. [DOI] [PubMed] [Google Scholar]
- 23.Li T, Chiang JY. Bile acid signaling in liver metabolism and diseases. Journal of Lipids. 2011b;2012:754067. doi: 10.1155/2012/754067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luo W, Friedman M, Shedden K, Hankenson K, Woolf P. GAGE: generally applicable gene set enrichment for pathway analysis. BMC Bioinformatics. 2009;10:161. doi: 10.1186/1471-2105-10-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McCullough AJ. Pathophysiology of nonalcoholic steatohepatitis. Journal of Clinical Gastroenterology. 2006;40 doi: 10.1097/01.mcg.0000168645.86658.22. [DOI] [PubMed] [Google Scholar]
- 26.Nagana GGA, Shanaiah N, Cooper A, Maluccio M, Raftery D. Bile acids conjugation in human bile is not random: new insights from 1H-NMR spectroscopy at 800-MHz. Lipids. 2009;44:527–535. doi: 10.1007/s11745-009-3296-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perez MJ, Briz O. Bile-acid-induced cell injury and protection. World Journal of Gastroenterology. 2009;15:1677–1689. doi: 10.3748/wjg.15.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ratziu V, de Ledinghen V, Oberti F, Mathurin P, Wartelle-Bladou C, Renou C, Sogni P, Maynard M, Larrey D, Serfaty L, Bonnefont-Rousselot D, Bastard JP, Riviere M, Spenard J. A randomized controlled trial of high-dose ursodeoxycholic acid for nonalcoholic steatohepatitis. Journal of Hepatology. 2011;54:1011–1019. doi: 10.1016/j.jhep.2010.08.030. [DOI] [PubMed] [Google Scholar]
- 29.Rezen T. The impact of cholesterol and its metabolites on drug metabolism. Expert Opinion on Drug Metabolism & Toxicology. 2011;7:387–398. doi: 10.1517/17425255.2011.558083. [DOI] [PubMed] [Google Scholar]
- 30.Russell DW. The enzymes, regulation, and genetics of bile acid synthesis. Annual Review of Biochemistry. 2003;72:137–174. doi: 10.1146/annurev.biochem.72.121801.161712. [DOI] [PubMed] [Google Scholar]
- 31.Sanyal AJ, Brunt EM, Kleiner DE, Kowdley KV, Chalasani N, Lavine JE, Ratziu V, McCullough A. Endpoints and clinical trial design for nonalcoholic steatohepatitis. Hepatology. 2011;54:344–353. doi: 10.1002/hep.24376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seyhun E, Malo A, Schaffer C, Moskaluk CA, Hoffmann RT, Goke B, Kubisch CH. Tauroursodeoxycholic acid reduces endoplasmic reticulum stress, acinar cell damage, and systemic inflammation in acute pancreatitis. American Journal of Physiology - Gastrointestinal and Liver Physiology. 2011;301:G773–G782. doi: 10.1152/ajpgi.00483.2010. [DOI] [PubMed] [Google Scholar]
- 33.Sharma R, Majer F, Peta VK, Wang J, Keaveney R, Kelleher D, Long A, Gilmer JF. Bile acid toxicity structure–activity relationships: Correlations between cell viability and lipophilicity in a panel of new and known bile acids using an oesophageal cell line (HET-1A) Bioorganic and Medicinal Chemistry. 2010;18:6886–6895. doi: 10.1016/j.bmc.2010.07.030. [DOI] [PubMed] [Google Scholar]
- 34.Song P, Zhang Y, Klaassen CD. Dose-response of five bile acids on serum and liver bile acid concentrations and hepatotoxicty in mice. Toxicological Sciences. 2011;123:359–367. doi: 10.1093/toxsci/kfr177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Svegliati-Baroni G, Ridolfi F, Hannivoort R, Saccomanno S, Homan M, de Minicis S, Jansen PLM, Candelaresi C, Benedetti A, Moshage H. Bile acids induce hepatic stellate cell proliferation via activation of the epidermal growth factor receptor. Gastroenterology. 2005;128:1042–1055. doi: 10.1053/j.gastro.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 36.Tan KP, Yang M, Ito S. Activation of Nuclear Factor (Erythroid-2 Like) Factor 2 by Toxic Bile Acids Provokes Adaptive Defense Responses to Enhance Cell Survival at the Emergence of Oxidative Stress. Molecular Pharmacology. 2007;72:1380–1390. doi: 10.1124/mol.107.039370. [DOI] [PubMed] [Google Scholar]
- 37.Thomas C, Pellicciari R, Pruzanski M, Auwerx J, Schoonjans K. Targeting bile-acid signalling for metabolic diseases. Nat Rev Drug Discov. 2008;7:678–693. doi: 10.1038/nrd2619. [DOI] [PubMed] [Google Scholar]
- 38.Timbrell JA, Seabra V, Waterfield CJ. The in vivo and in vitro protective properties of taurine. General Pharmacology: The Vascular System. 1995;26:453–462. doi: 10.1016/0306-3623(94)00203-y. [DOI] [PubMed] [Google Scholar]
- 39.Trauner M, Claudel T, Fickert P, Moustafa T, Wagner M. Bile acids as regulators of hepatic lipid and glucose metabolism. [Review] [14 refs] Digestive Diseases. 2010;28:220–224. doi: 10.1159/000282091. [DOI] [PubMed] [Google Scholar]
- 40.Trauner M, Fickert P, Halilbasic E, Moustafa T. Lessons from the toxic bile concept for the pathogenesis and treatment of cholestatic liver diseases. Wien Med Wochenschr. 2008;158:542–548. doi: 10.1007/s10354-008-0592-1. [DOI] [PubMed] [Google Scholar]
- 41.Trottier J, Bialek A, Caron P, Straka RJ, Heathcote J, Milkiewicz P, Barbier O. Metabolomic profiling of 17 bile acids in serum from patients with primary biliary cirrhosis and primary sclerosing cholangitis: A pilot study. Digestive and Liver Disease. 2012;44:303–310. doi: 10.1016/j.dld.2011.10.025. [DOI] [PubMed] [Google Scholar]
- 42.Trottier J, Bialek A, Caron P, Straka RJ, Milkiewicz P, Barbier O. Profiling circulating and urinary bile acids in patients with biliary obstruction before and after biliary stenting. PLoS ONE. 2011;6:e22094. doi: 10.1371/journal.pone.0022094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang JS, Kim JT, Jeon J, Park HS, Kang GH, Park KS, Lee HK, Kim S, Cho YM. Changes in hepatic gene expression upon oral administration of taurine-conjugated ursodeoxycholic acid in ob/ob mice. PLoS ONE. 2010;5:e13858. doi: 10.1371/journal.pone.0013858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Y, Hong JY, Rockwell CE, Copple BL, Jaeschke H, Klaassen CD. Effect of bile duct ligation on bile acid composition in mouse serum and liver. Liver International. 2012;32:58–69. doi: 10.1111/j.1478-3231.2011.02662.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zollner G, Wagner M, Fickert P, Silbert D, Gumhold J, Zatloukal K, Denk H, Trauner M. Expression of bile acid synthesis and detoxification enzymes and the alternative bile acid efflux pump MRP4 in patients with primary biliary cirrhosis. Liver International. 2007;27:920–929. doi: 10.1111/j.1478-3231.2007.01506.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.