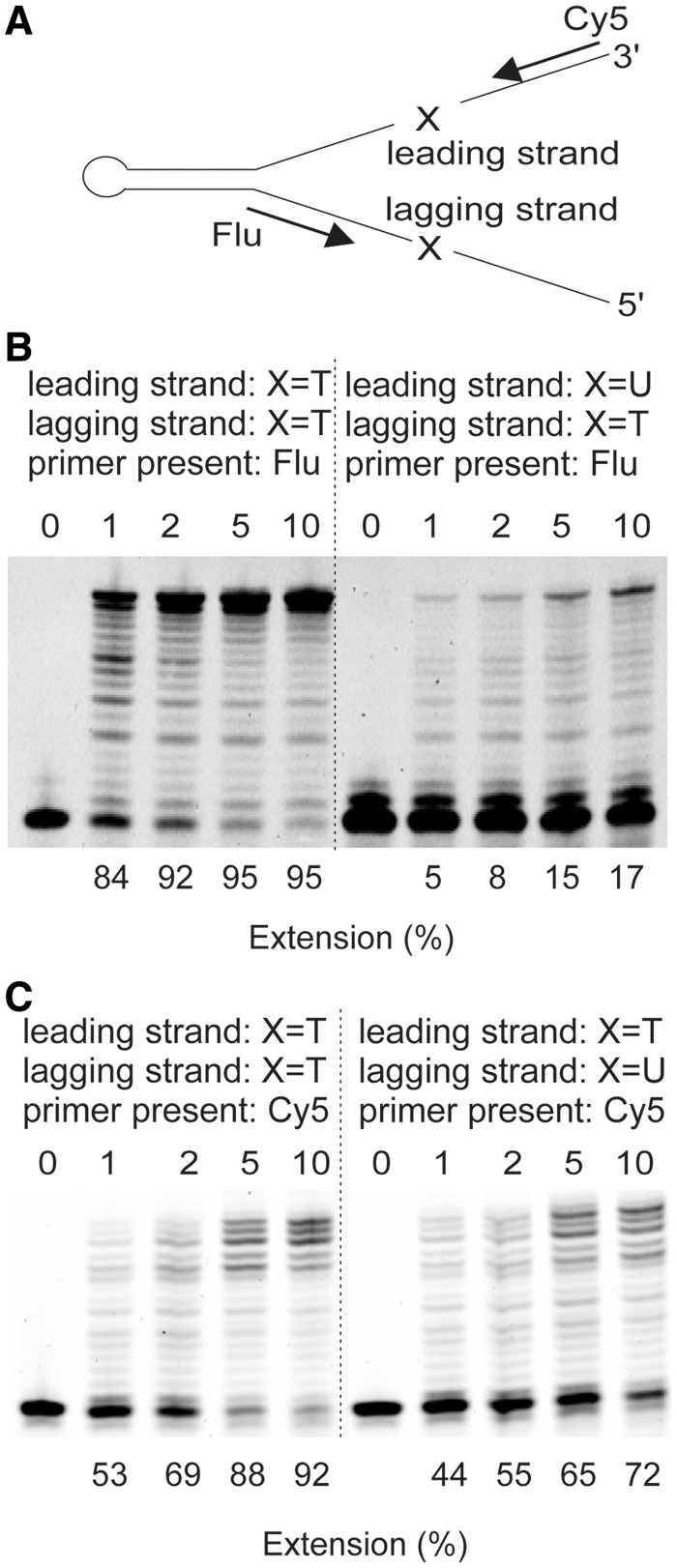
Figure 4.
Inhibition of Pfu-Pol D by ‘trans’ located uracil. (A) Structure of the primer-template mimic (for full sequences, see Supplementary Figure S4). A long ‘snap-back’ oligodeoxynucleotide forms the backbone of the mimic. Annealing of primers (either singly or in pairs) produces leading and lagging strand branches which can contain a single uracil residue (indicated by X) four bases ahead of the primer-template junction. (B) Results seen when the lagging strand is copied (only Flu primer present) with uracil (thymine in controls) present in the leading strand. (C) Results seen when the leading strand is copied (only Cy5 primer present) with uracil (thymine in controls) present in the lagging strand. Here, only one primer was used per experiment. For the results observed using two primers for each experiment, see Supplementary Figure S4.