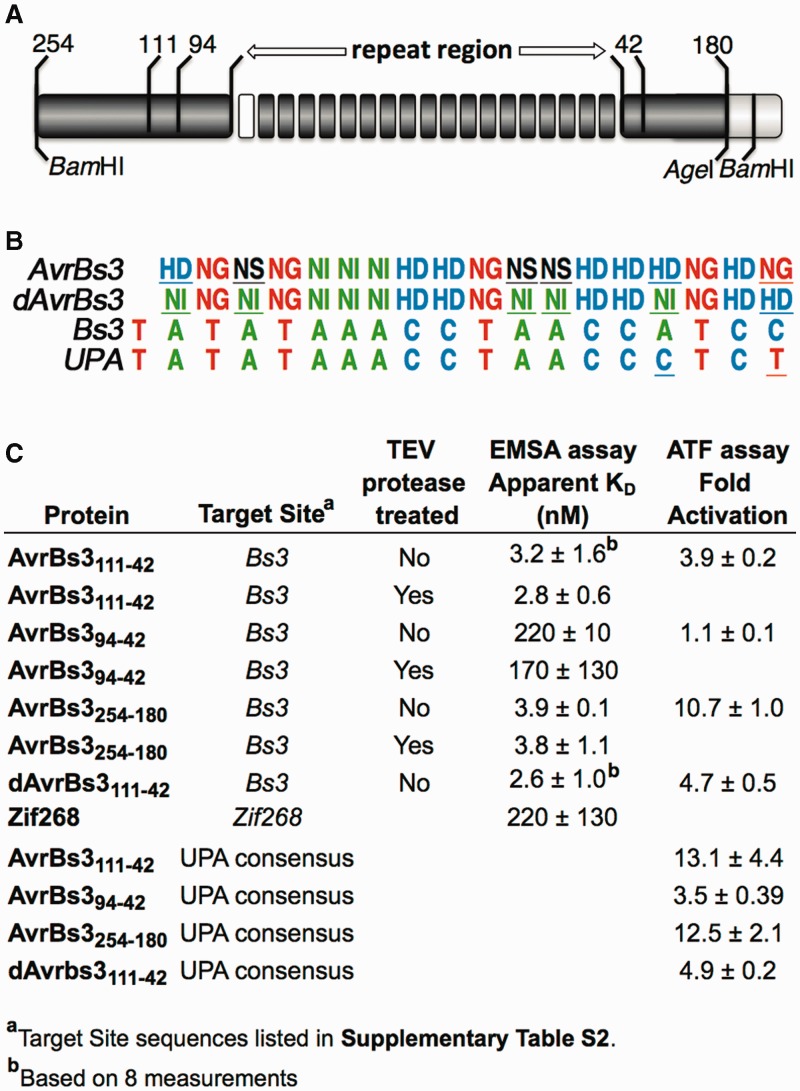
Figure 1.
Affinity and transcriptional activation data for several AvrBs3 variants. (A) Schematic of a TALE polypeptide showing the 18 RVD-containing repeats with N- and C-terminal flanking regions. The ‘0 repeat’ is shown in white. The numbers indicate the lengths of the N- and C-terminal extensions outside the repeat region used in the different constructs described in this work. A comprehensive survey of N- and C-terminal boundaries used in previous TALE studies is given in Supplementary Figure S1. (B) RVD amino acid composition of AvrBs3 (first row), along with the sequence of a natural DNA target, Bs3 (third row), and the consensus AvrBs3 site, UPA (2) (fourth row). The RVD composition of the dAvrBs3 variant, which contains only the standard NI, HD and NG RVDs and no mismatches to the Bs3 box target site, is also shown (second row). AvrBs3 RVDs that are ‘non-standard’ or mismatched to Bs3, and the corresponding RVDs in dAvrBs3, are underlined. The UPA site bases that differ from Bs3 are also underlined. (C) EMSA and ATF activation data were obtained as described in Materials and Methods. Target site sequences and RVD compositions are listed in Supplementary Tables S2 and S3. The affinity of Zif268 was measured in TALE 1× binding buffer. Zif268 affinity measured in a standard zinc-finger binding buffer (16) was more typical, 11 ± 4 nM.