Abstract
ZNF143 is a zinc-finger protein involved in the transcriptional regulation of both coding and non-coding genes from polymerase II and III promoters. Our study deciphers the genome-wide regulatory role of ZNF143 in relation with the two previously unrelated transcription factors Notch1/ICN1 and thanatos-associated protein 11 (THAP11) in several human and murine cells. We show that two distinct motifs, SBS1 and SBS2, are associated to ZNF143-binding events in promoters of >3000 genes. Without co-occupation, these sites are also bound by Notch1/ICN1 in T-lymphoblastic leukaemia cells as well as by THAP11, a factor involved in self-renewal of embryonic stem cells. We present evidence that ICN1 binding overlaps with ZNF143 binding events at the SBS1 and SBS2 motifs, whereas the overlap occurs only at SBS2 for THAP11. We demonstrate that the three factors modulate expression of common target genes through the mutually exclusive occupation of overlapping binding sites. The model we propose predicts that the binding competition between the three factors controls biological processes such as rapid cell growth of both neoplastic and stem cells. Overall, our study establishes a novel relationship between ZNF143, THAP11 and ICN1 and reveals important insights into ZNF143-mediated gene regulation.
INTRODUCTION
The transcriptional regulatory system plays a fundamental role in controlling the correct expression of genes involved in many biological processes (1). This mechanism involves specific DNA-binding transcription factors, co-factors and chromatin remodelling factors. In humans, there are nearly 1400 transcription factors of which only a few have been extensively studied (1). They bind to cis-regulatory elements located in the promoters of specific genes and modulate their activation or repression (2). Therefore, their influence on gene expression is achieved by acting directly, or via a multiplicity of partners, on chromatin remodelling and recruitment of the transcription machinery (3). Despite our limited knowledge of the function of all transcription factors, cis-regulatory regions of genes are broadly studied nowadays at the genome-wide scale (4). Owing to the increasing number of high-throughput studies (5,6), numerous transcription factors are found to be located within the same promoter regions affecting or not the gene expression, acting in synergy or having antagonistic effects depending on physiological and environmental conditions (7,8). Thereby combinatorial binding of transcription factors modulates gene expression according to the needs and conditions of cells (9).
In this regard, we were first interested in deciphering the genome-wide regulatory potential of the transcription factor ZNF143. Also known as Staf (Seleno cysteine tRNA gene transcription activating factor), it is a zinc-finger protein involved in the control of both coding and non-coding genes from RNA polymerase II (Pol II) and RNA polymerase III (Pol III) promoters (10). This factor recognizes and binds with high affinity a well-characterized 18-bp motif located in the core promoter region. ZNF143 has been shown to be involved in the activation of several ncRNA (10–12) and protein-coding genes (13–15). It is conserved in all chordates, and its vertebrate paralogue ZNF76 (16) possesses an identical central 209 amino acid long DNA-binding domain (DBD). ZNF143 is involved in the resistance to cis-platin in cancer cells through the transcriptional regulation of DNA repair genes (17). Moreover, this factor is critical for the normal development of zebrafish embryo (18). ZNF143 and Zfp143, the murine orthologue, are important for the maintenance of pluripotency of embryonic stem cells (19,20). Considering this evidence, it is likely that ZNF143 plays a role in the regulation of gene expression in rapidly growing cells.
ICN1 is the active form of the Notch1 receptor, which activates transcription of target genes through binding to the DNA-binding repressor RBPJ (21). In the canonical Notch1 pathway, after binding of the ligand (DELTA and JAGGED family members), the Notch1 receptor is cleaved through highly regulated step-wise processes carried out by the γ-secretase (21,22). The released active intracellular form ICN1 enters the nucleus and forms a transcriptional complex with RBPJ that recognizes a 7-bp motif TTCCCAA on the DNA (23). Although the canonical Notch1 pathway, mediated by cell–to-cell communication, is implicated in diverse developmental biological processes (24), the aberrant and increased activity of Notch1 signalling is present in many human cancers and implicated in regulating self-renewal of stem cell-like cells in tumours (22). One of the most notable disorders mediated by the deregulated Notch1 pathway is the T-Acute Lymphoblastic Leukaemia (T-ALL), in which gain-of-function mutations in Notch1 lead to ligand-independent ICN1 overproduction in leukaemic blasts (23).
Thanatos-associated protein 11 (THAP11) is a C2CH zinc-finger transcription factor, binding specifically a 15-bp long bipartite DNA motif (25). This ubiquitous protein is overexpressed in colon cancer cells (26) and in embryonic stem cells (27), where it plays a specific role in cell proliferation. Until recently, it was admitted that THAP11 had a repressor function mediated by its co-factor HCF-1 (27). However, it was recently shown that for some genes, THAP11/HCF1 could act as a transcription activator as well (25,26). In mouse, Ronin (THAP11 orthologue) is essential for embryogenesis and the pluripotency of embryonic stem cells (mES cells) (27). Recent work showed that Ronin-targeted genes in mouse mES cells function in protein biosynthesis and energy production, suggesting a major role in cell growth (25).
In this work, we report the genome-wide association to DNA of ZNF143, ICN1 and THAP11 transcription factors in different human and murine cells. Owing to similarities in target genes and DNA-binding motifs, we also investigated their regulatory potential by combining transcriptome analysis with chromatin immunoprecipitation followed by high-throughput sequencing (ChIP-Seq) data. This study presents strong evidence that the three factors play a pivotal role in modulating expression of a common set of genes controlling biosynthetic pathways via the mutually exclusive occupancy of overlapping binding sites.
MATERIALS AND METHODS
Antibodies and constructs
Antibodies and constructs used in these studies are described in Supplementary Materials and Methods (Supplementary data).
Generation of stable cell lines and induced protein expression
Flp-InTM T-RexTM 293 system (Invitrogen) was used according to the manufacturer’s instructions to generate stable cell lines with regulated expression of ZNF143 (FLP143), ZNF143-3xHA (FLP143-HA), ZNF76 (FLP76) RBPJ-3xHA (FLP_RBPJ-HA) and THAP11-3xHA (FLPTHAP11-HA). Stable clones containing the required open reading frame under the control of the cytomegalovirus/tetO2 hybrid promoter were selected by culturing in selective medium containing 250 µg/ml of hygromycin (InvivoGen) and 15 µg/ml of blasticidin (InvivoGen). Resistant colonies were expanded and tested for doxycycline-regulated protein expression. Protein expression was induced by addition of 1 µg/ml of doxycycline in culture medium.
ChIP and ChIP-Seq
ChIP assays were performed essentially as previously described (28) with 1–4 × 107 cells. Chromatin was sonicated to an average of 200 bp using a BioruptorTM UCD-200 (Diagenode) at maximum power level for 30 min (pulses of 30 s with 30 s breaks). After DNA purification, enrichment was estimated by quantitative real time PCR (qPCR). qPCR was performed in duplicate on a Stratagene Mx3005P PCR system (Agilent Technologies) using 5× HOT Pol® EvaGreen®qPCR Mix PLUS (ROX) (Euromedex) and input DNA as the standard. Promoters containing, and regions devoid of the studied binding sites, were used as positive and negative controls, respectively. Enrichment was determined by the ΔΔCt method. Primer sequences are available on request. Samples were sequenced using the Illumina Genome Analyzer GAII, and the read sequences were mapped to unmasked version of the human (hg19) or mouse (mm9) genomes via the Illumina Eland V1.6 (Illumina) or by the Bowtie software (29) with either an exact 36 nt match or with a single or two mismatches. Reads mapped to multiple locations were discarded, and no more than two redundant reads having exactly the same genomic coordinates were used to identify enriched regions. Tag mapping in the large families of highly related ncRNA genes was performed with the Bowtie software, allowing two perfect matches in the genome. ChIP-Seq data were submitted to the Gene Expression Omnibus database and assigned the identifier GSE39263.
Description of ChIP-Seq data processing and integrative analysis is available in Supplementary Materials and Methods (Supplementary data).
Electrophoretic mobility shift assay
Recombinant ZNF143 DBD, RBPJ from VK91 (30) and THAP11 DBD were produced using the GST gene fusion system as previously described (15,30) essentially according to the manufacturer’s instructions (GE Healthcare). Dissociation constants of ZNF143 on the SBS1 (GTTATGGAATTCCCATTATGCACCGCG) or SBS2 (AAACTACAATTCCCATTATGCACCGCG) motifs were derived as previously described (16). Internally labelled DNA fragments containing the SBS1 (TTCCCATTATGCACCGCG), SBS2 (ACTACAATTCCCATTATGCACCGCG), THAP11-binding site (TBS) (ACTACAATTCCCA) and ACTACAA motifs were produced by PCR in the presence of 50 µCi of (α-32P)dCTP (3000 Ci/mmol). Binding assays were performed essentially as described (15) with 20 fmol of labelled probe. Protein–DNA complexes were resolved by electrophoresis on 4% native polyacrylamide gels containing 0.25 × Tris-borate-EDTA. Competition was performed with a 200-fold molar excess of unlabelled specific competitor or unspecific competitor.
RNA preparation and expression analysis
ZNF143 knockdown was performed in HeLa cells as described previously (28). The THAP11-specific Mission esiRNA (Sigma-Aldrich) targeting positions 872–1276 of the THAP11 mRNA or control, esiRNA targeting GFP (Sigma-Aldrich) were transfected into HeLa cells using Lipofectamine2000 according to manufacturer’s instructions (Invitrogen). For γ-secretase inhibition, HPB-ALL cells were treated with 500 nM of Compound E (Enzo Life Sciences), and control was obtained with DMSO-treated HBP-ALL cells. ZNF143-HA or ZNF143 overexpression was induced by treatment of FLP143-HA or FLP143 cells with 1 µg/ml of doxycycline, and cells were collected at different time points post-induction. Total RNA was extracted using TRIREAGENT (Euromedex) and reverse transcribed with dN9 oligonucleotides. The complementary DNA obtained was amplified with specific primers on the Stratagene Mx3005P PCR system (Agilent Technologies) using EvaGreen qPCR Mix Plus (Euromedex). Primer sequences are available on request. All reactions were carried out in triplicate. The relative expression ratio was calculated using the REST 2009 software (Qiagen).
RNA-seq differential expression analysis
Total RNA was isolated from HeLa-S3 and FLP143 cells treated as described earlier in the text. After selection of polyA + mRNA, RNA-seq libraries preparation was performed using Illumina TruSeq RNA Sample Preparation kit according to manufacturer protocol. All RNA samples were sequenced using the Illumina Genome Analyzer GAII and aligned to the hg19 reference genome using TopHat software (31). Aligned reads were processed with Cuffdiff (32) to measure the differential expression using the RefSeq genes as reference annotation. Only the transcripts matching the genes potentially regulated by ZNF143 (TSS located at +/−2 kb of a ChIP-Seq peak summit) were analysed, and the expression value for each transcript was measured by a FPKM value (Fragments per kilobase of transcripts per million mapped tags). RNA-seq data were submitted to the Gene Expression Omnibus database and assigned the identifier GSE39263.
Data access
Data from this study were submitted to the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under accession number GSE39263.
RESULTS
Genome-wide identification, characterization and distribution of ZNF143-binding events in different human and mouse cell lines
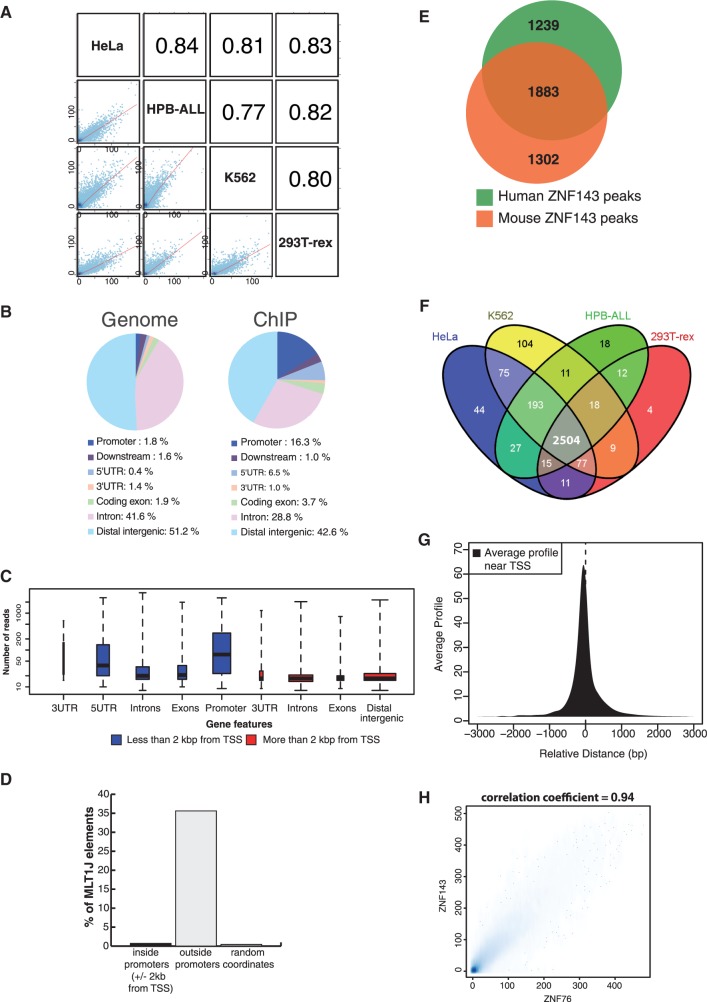
To identify ZNF143-binding events, ChIP-Seq was performed with anti-ZNF143 antibody in four human (HeLa-S3, T-REx HEK293, K562 and HPB-ALL; Supplementary Figure S1A) and three mouse cell types (NIH3T3, mESC and MEF; Supplementary Figure S1B). Quantitative enrichment analysis of the ZNF143 peaks showed a strong correlation (coefficient between 0.77 and 0.93) between the experiments (Figure 1A and Supplementary Figure S2). The locations of ZNF143-binding events show an important enrichment in peaks located at +/−2 kb from the transcription start site (TSS) of protein coding genes (50 and 59% of all human and mouse ZNF143-binding events, respectively) (Supplementary Table S1) (Figure 1B and C and Supplementary Figure S3A). In human and mouse cells, we identified the MLT1J retrotransposon subfamily, showing a significant enrichment in ZNF143 peaks compared with random coordinates (88.9-fold enrichment in HeLa cells). MLT1J retroelements associated with ZNF143 are essentially located outside of promoters and represent 35.5% of repeat elements overlapping ZNF143-binding events in HeLa cells (Figure 1D). We will focus now on ZNF143-binding events located and enriched at proximity of the TSS of coding genes (+/−2 kb from a TSS) (Supplementary Figure S3B). We identified, at these locations, 3122 ZNF143-binding events in the set of human (Supplementary Table S4) and 3385 in the set of mouse cells (Supplementary Table S5), 60.3% of which being conserved in humans (Figure 1E). We observed a core set of 2504 ZNF143-binding regions shared by the four human cell lines (Figure 1F) and one of 2686 shared by the three mouse cell types (data not shown). The binding events were essentially present in the 500 bp upstream of the TSS (Supplementary Figure S4) with a positional preference at 70 bp upstream of it (Figure 1G). We observed also an important enrichment of the ZNF143-binding events located in bidirectional promoters. In human cells, 661 binding events are located within bidirectional promoters (Supplementary Table S4), representing 17% of all the bidirectional promoters in the genome. The chromosomal distribution of the ZNF143-binding events showed a noteworthy enrichment of the ZNF143 peaks in promoters of genes encoding krüppel-associated box (KRAB) zinc-finger proteins on the human chromosome 19 and mouse chromosome 7 (Supplementary Figure S5). Finally, we examined the ZNF143 occupancy in five sets of ncRNA genes (RefSeq ncRNA, ensembl ncRNA, lincRNA, TUCP and Pol III ncRNA). We identified 476 ZNF143-binding events located at +/−2 kb from the TSS of a ncRNA gene belonging to four subsets (Supplementary Table S6 and Supplementary Figure S6). Overall, subset 1 includes binding events at proximity of lincRNA, TUCP and other ncRNA genes. Subset 2 includes binding events at proximity of tRNA, miRNA and snoRNA/scaRNA genes. The latter two subsets 3 and 4 contain Pol III type 3 ncRNA genes and Pol II snRNA genes, respectively (Supplementary Table S7).
Figure 1.
Genome-wide characterization of ZNF143-binding events in human and mouse cells. (A) Correlation plots of ZNF143-binding events in four human cell lines. Grid of pairwise scatter plots with associated Pearson correlation coefficient obtained using Cistrome software (33). The blue dots coordinates represent the normalized enrichments in reads for each cell lines at all the peaks coordinates (Supplementary Table S1). The regression line is indicated in red and the Pearson correlation coefficients are indicated in the squares on the top right (B) Pie charts showing distribution of the ZNF143-binding events in different genomic regions. A promoter is defined as a region within −2 kb from the TSS of a gene; downstream, 2 kb downstream of the end of a gene; the distal intergenic are regions excluding a promoter, downstream, 5′UTR, 3'UTR, introns and exons. Distribution of the different genomic regions in the human genome is given as a reference. Analysis was performed using the coordinates of the peaks summit in HeLa-S3 cells (Supplementary Table S1, P-value threshold of 10−5) expanded to 400 bp (+/−200 bp on both sides of the summit). (C) Box plots depicting the read enrichment of peaks located in various genomic regions. The peaks and enrichment value (in number of reads) are from the HeLa-S3 ChIP-Seq experiment. The blue and red box plots correspond to genomic regions located at less and more than 2 kb from a TSS, respectively. The size of each population of peaks is reflected by the width of the plot. (D) Distribution of MLT1J elements in ZNF143 peaks located inside (+/−2 kb from a TSS) and outside of promoters. The random coordinates were generated, 10 times, by random permutation of the set of peak coordinates located outside promoters across the genome. MLT1J elements are expressed in percentage of MLT1J in repeat elements that overlap ZNF143-binding events. (E) Venn diagram depicting the overlap of ZNF143-binding events located inside promoters in mouse and human cells, respectively. The analysis was performed using the complete data set listed in Supplementary Tables S4 and S5 with coordinate conversion from mm9 to hg19. (F) Venn diagram showing the overlap between the ZNF143-binding events identified at +/−2 kb from a TSS (Supplementary Table S4) in HeLa-S3, K562, 293T-Rex and HPB-ALL human cells. (G) Positional preference of the ZNF143-binding events in promoters. Average profile of the 3122 human ZNF143-binding events (Supplementary Table S4) located at +/−2 kb from a TSS. Positions are given in nucleotides (nt), and negative values correspond to locations upstream from the TSS. (H) Comparison between all the ZNF143- and ZNF76-binding events identified by ChIP-Seq in stable cell lines expressing ZNF143-HA or ZNF76. The scatter plot represents the enrichment correlation of both experiments (Supplementary Table S1). Scale represents normalized number of reads.
Two distinct motifs with overlapping sequences are associated to the in vivo ZNF143-binding events
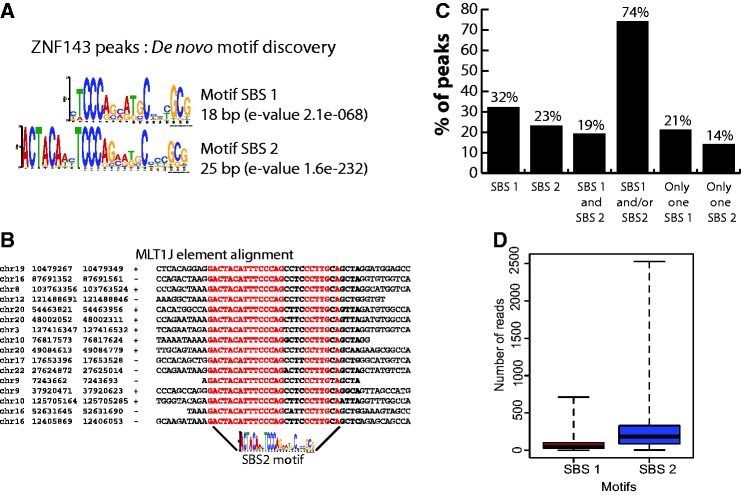
We identified by a motif search conducted on all ZNF143-binding events the known 18-bp ZNF143-binding motif (10) and a second 25-bp motif containing the first one with an additional ACTACAN sequence located immediately upstream (Figure 2A). Henceforth, we will refer to the canonical 18-bp motif as SBS1 and the 25-bp motif as SBS2 (Figure 2A). The ZNF143-binding events that overlap MLT1J retroelements contain only the SBS2 motif (Figure 2B). For motifs associated to the ZNF143 peaks identified at proximity of ncRNA genes, we determined that the binding events in subsets 1 and 2 (Supplementary Figure S6) include the SBS1 and/or SBS2 motifs. Surprisingly, all the 56 ZNF143-binding events associated to Pol III type 3 ncRNA and Pol II snRNA genes (subsets 3 and 4 in Supplementary Figure S6) contain only one SBS1 motif while we would expect only 10 in respect of the distribution of SBS1 and SBS2 motifs in the all binding events (z-test: P < 10−5). Subset 3 contains peaks located at proximity of the TRNAU1 (tRNASec), RNU6.1, RNU6.2, RNAU6.7-U6.9, RN7SK, RPPH1, RNY1, RNY3, RNY4 and RNY5 genes (Supplementary Table S7). Supplementary Figure S7A (lower part) displays a multiple alignment showing sequences of the four elements (TATA element, PSE promoter element, octamer motif and SBS1) (35) constituting the promoters of the 12 genes cited earlier in the text. The position of the SBS1 motif is restricted to a window covering positions −245/−199, except for the promoter of the RPPH1 gene (11) where SBS1 is located at position −87. Subset 4 (Pol II snRNA genes) contains the RNU1, RNU2, RNU4, RNU4ATAC, RNU5, RNU8, RNU11 and RNU12 genes (Supplementary Table S7). The multiple sequence alignment in Supplementary Figures S7A (upper part) and S7B shows the presence of canonical PSE and octamer motifs at proximity of SBS1 in 41 of the 44 Pol II snRNA genes (Supplementary Table S7). We will henceforth focus on the SBS1 and SBS2 motifs identified in promoters (+/−2 kb from a TSS) of protein coding genes in humans. The SBS1 and/or SBS2 motifs were detected in 74% of the 3122 binding events (Figure 2C and Supplementary Table S4). As ZNF143 could be targeted to certain locations by some of its putative partners, we searched for known binding sites in the peaks without any obvious ZNF143-binding motif. The only other motif reported was an Sp1-like binding site; it was enriched around the peaks, reflecting per se the association of ZNF143 with CpG islands. Figure 2C shows that the family of peaks with SBS1 and/or SBS2 motifs consists of three subfamilies: one subfamily (32% of all peaks) contains only SBS1 motifs, the second (23%) only SBS2 motifs and the third one (19%) both motifs. We observed that 21 and 14% of all binding events contain only a single SBS1 or SBS2 motif, respectively (Figure 2C). Strikingly, in contrast to the binding events with only an SBS1 or an increasing number of SBS1 motif, the ZNF143-binding events with a single or increasing number of SBS2 motifs showed a increasing ChIP score (t-test: P < 10−5) (Figure 2D and Supplementary Figure S8) Finally, we examined the functional annotation of genes at proximity of the 2504 core set of ZNF143-binding events shared by the four human cell lines (Figure 3A). We compared then four sets of genes identified at proximity of the peaks and containing different combinations of motifs (SBS1 motif only, SBS2 motif only, both motifs) or without motif (Supplementary Figure S9). Overall, the functional analysis revealed that all the sets of genes are enriched in terms associated to cell growth and primary metabolism. Nevertheless, a high enrichment score was found for terms such as translation for the set of genes associated to SBS1 and transcription regulation and zinc fingers for the SBS2 motif. To attest the involvement of ZNF143 in cell growth and primary metabolism, we tested the impact of ZNF143 knockdown on HeLa-S3 cells proliferation. As expected and as previously shown for PC3 cells (37), HeLa-S3 cells had a significantly reduced proliferation rate at 72 h post-transfection with siRNA targeting specifically ZNF143 versus siRNA-targeting GFP (Figure 3B).
Figure 2.
Identification and characterization of ZNF143-binding motifs. (A) Sequence logo depicting the SBS1 and SBS2 motifs discovered de novo at +/−100 bp ZNF143 peak summits in human and mouse genomes, using the MEME suite (34). (B) Multiple sequence alignment depicting some of the MLTIJ elements that are bound by ZNF143. Coordinates of the repeated elements are shown on the left with indication of the orientation of the element (+/−). (C) ZNF143-binding event counts in human genome associated with SBS1, SBS2, SBS1 and SBS2, a single SBS1 or a single SBS2 motif. (D) Box blots showing ChIP-Seq score (number of reads) distribution of ZNF143-binding events containing: only SBS1 or SBS2 motifs.
Figure 3.
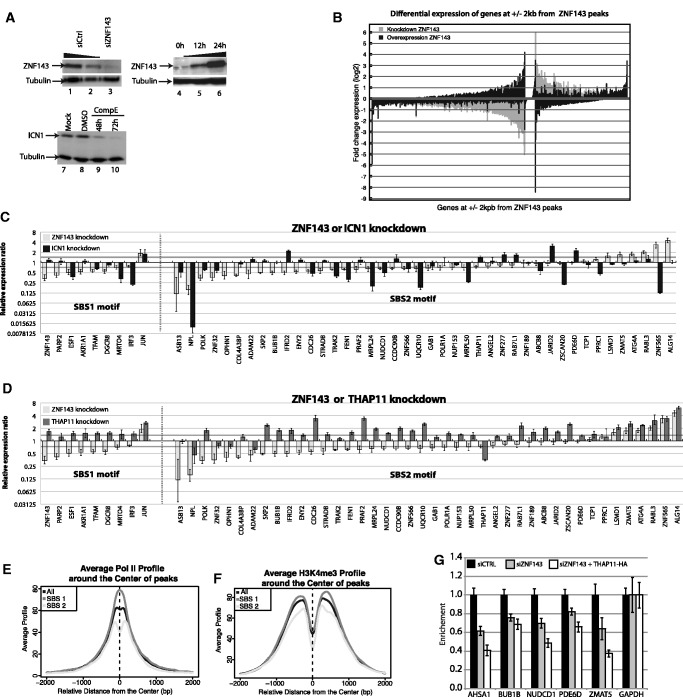
ZNF143-targeted genes are involved in cell growth and proliferation. (A) Functional categories enriched among the genes located at +/−2 kb of the identified ZNF143-binding events common to HeLa-S3, K562, T-Rex-293 and HPB-ALL cell line (Supplementary Table S4), as reported by the DAVID web-based functional annotation program (36). Values are fold-enrichment scores compared with the whole set of human genes used as background. (B) Cell proliferation assay performed for a 72 h time course on HeLa-S3 cells non-transfected (no siRNA) or transfected with siRNA-targeting ZNF143 or GFP. Results are expressed as means +/− standard deviation of three biological replicates.
Genome-wide analysis of ZNF76 binding in T-REx HEK293 cells reveals a total overlap with ZNF143 binding
In mammalian cells, ZNF76 was characterized as the ZNF143 paralogue with an activator and repressor function (16,38). We asked whether the identified ZNF143 targets are recognized by ZNF76 in human T-Rex-HEK293 cells expressing ZNF76 (Supplementary Figure S1D). To facilitate ChIP, clonal cell lines expressing ZNF76 were generated (Supplementary Figure S1A), and we characterized the loci occupied by this factor by ChIP-Seq. The data were compared with those obtained by the anti-HA ChIP-Seq on FLP143-HA cells. An enrichment correlation coefficient of 0.94 was observed between the two experiments for all of the binding events (Figure 1H). Obviously, a de novo motif search showed that the ZNF76-binding events include the SBS1 or SBS2 motifs described earlier in the text. Moreover, both ZNF76 and ZNF143 behave, in terms of enrichment on SBS1 or SBS2 sites, in an identical way. This is illustrated in Supplementary Figure S10 with the peaks formed on the ATP5J2 (SBS1 only) and NUP153 (SBS2 only) promoters. We evidenced also that ZNF76 is associated to ncRNA genes with a total overlap with the ZNF143-binding events.
THAP11 and ZNF143 bind genome-wide to SBS2 sites through mutually exclusive events
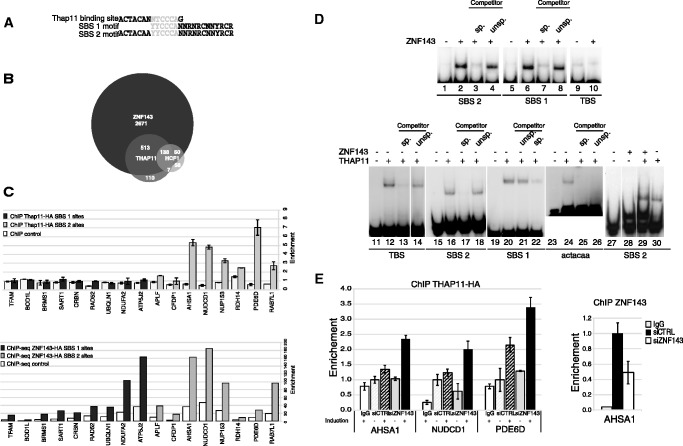
The widespread association of SBS1 and/or SBS2 motifs to ZNF143-binding events led us to examine whether the additional ACTACAN sequence in SBS2 affects ZNF143 binding. The apparent Kd of ZNF143 for the SBS1 (1.36 +/−0.33 nM) and SBS2 (1.58 +/−0.19 nM) motifs are very similar (Supplementary Figure S11), suggesting that the 5′ part of the SBS2 motif (ACTACAN), or a more extended sub-motif, constitutes a binding site for another factor. In this respect, ChIP-Seq on mES cells revealed that the transcription factor Thap11 binds with its transcriptional co-regulator Hcf-1 to the sequence motif ACTACANNTCCCA (TBS) (Figure 4A) (25), suggesting a possible overlap of ZNF143 and Thap11 DNA-binding events. We compared the available ChIP-Seq data of Thap11 and its co-factor Hcf-1 (25) obtained from mES cells with our ZNF143 ChIP-Seq data obtained from the same cells. We found that 84.7% of all Thap11 and 75.8% of all Hcf-1-binding events located in promoters overlap the ZNF143-binding events located at +/−2 kb of a TSS (3385 peaks) (Figure 4B). An individual motif occurrence search on the 651 peaks, common to the Thap11- and ZNF143-binding events (Figure 4B), identified an SBS2 motif in the whole set of coordinates. Furthermore, a similar search performed on Hcf-1 peaks without reported Thap11 binding, showed that 51% (Figure 4B) of them overlap with ZNF143 and contain also an SBS2 motif. This suggests that Thap11 could in fact also bind these SBS2 sites. From these observations, it appears that Thap11 binding is restricted to loci containing an SBS2 motif. By ChIP, we asked whether the promoters containing only SBS1 (SBS1 loci) or SBS2 (SBS2 loci) motifs are recognized by THAP11. We generated clonal cell lines expressing HA-Tagged THAP11 (FLPTHAP11-HA) and by anti-HA ChIP, we examined and compared the THAP11 and ZNF143 occupancy on SBS1 and SBS2 loci (Figure 4C). As expected, ChIP-Seq showed ZNF143 occupancy on the nine SBS1 loci and on the eight SBS2 loci (Figure 4C, lower part). In contrast, THAP11 occupancy was restricted to the SBS2 loci without significant occupation of the SBS1 loci (Figure 4C, upper part). By gel-shift experiments, we further examined the in vitro binding of ZNF143 and THAP11 DBDs on TBS, SBS1 and SBS2 motifs. As expected, the ZNF143-DBD binds specifically to the SBS1 and SBS2 motifs (Figure 4D, lanes 1–8) but is unable to recognize the TBS (Figure 4D, lanes 9 and 10). THAP11–DBD recognized specifically the TBS (Figure 4D, lanes 11–14) and the SBS2 motif (Figure 4D, lanes 15–18), which per se contains a TBS. Surprisingly, unlike the TBS sub-motif ACTACAA (Figure 4D, lanes 23–26), the SBS1 motif, which contains only the 3′ part of TBS (TTCCCA) binds specifically the THAP11–DBD (Figure 4D, lanes 19–22). This observation suggests that THAP11 has a higher affinity to the 3′-part of the TBS (TTCCCAN). Binding assays performed on the SBS2 motif in the presence of ZNF143-DBD and THAP11–DBD revealed that the binding of the two proteins is mutually exclusive (Figure 4D, lanes 27–30). Indeed, the shift observed in the presence of THAP11 (Figure 4D, lane 29) is not altered by the presence of ZNF143–DBD (Figure 4D, lane 30). We also tested in vivo the THAP11 occupancy by ChIP on loci containing SBS2 motifs after ZNF143 siRNA-mediated knockdown in non-induced or induced FLPTHAP11-HA cells. The induced expression of THAP11 increases the occupancy of the factor on the promoters (Figure 4E, left part, compare induced and non-induced after siCTRL treatment). This phenomenon is amplified after THAP11 induction performed on cells with efficient ZNF143 siRNA-mediated knockdown (Figure 4E, left part, compare induced and non-induced after siRNA ZNF143). The efficiency of ZNF143 depletion was validated by anti-ZNF143 ChIP qPCR, as illustrated in Figure 4E (right part) for the AHSA1 promoter.
Figure 4.
Overlapping of ZNF143 and Thap11 DNA-binding events. (A) Sequence alignment of the TBS, SBS1 and SBS2 motifs. The sequence indicated in light grey is shared by the three motifs. (B) Venn diagram depicting the overlap of ZNF143/Zfp143-, Thap11- and Hcf1-binding events in mouse ES cells. The analysis was performed using the complete data set listed in Supplementary Table S5 and published ChIP-Seq data (25). (C) Histograms depicting the enrichment of genomic regions containing only SBS1 or SBS2 motifs in ChIP samples. ChIP assays with anti-HA antibody were performed on induced FLP143-HA and FLPTHAP11-HA stable cell lines. Uninduced stable cell lines were used as control. THAP11-HA data are obtained from ChIP-qPCR experiment, amplifying a promoter region containing the binding site. Primer sequences are available on request. ZNF143-HA data are obtained from ChIP-Seq experiment, using read number values in regions amplified by qPCR for THAP11-HA, expanded to 200 bp. SBS1 sites and SBS2 sites correspond to promoters containing only the SBS1 motif or only the SBS2 motif, respectively. Fold enrichment in promoters was normalized to control regions located at 2 kb of tested promoters. The gene symbols listed are those of the genes closest to the tested genomic region. (D) ZNF143 and THAP11 in vitro binding assays on SBS1, SBS2, TBS and TBS sub-motif. Gel retardation assays were performed with 32P labelled double-stranded fragment containing the indicated motif. The labelled probe was incubated in the presence (+) or absence (−) of ZNF143 or THAP11 DBD. The reactions in lanes 3, 7, 13, 17, 22, 25 and 4, 8, 14, 18, 21 were performed in the presence of an excess of unlabelled specific (sp.) or unspecific (unsp.) competitors. The ACTACAA probe corresponds to the 5′part of the TBS. Each lane presented in a single panel of the gel picture was from the same gel and the same exposure of the autoradiogram. (E) Histograms depicting the enrichment of promoter regions containing SBS2 motifs in ChIP samples. ChIP assays with anti-HA antibody were performed on induced or non-induced FLPTHAP11-HA stable cell line, transfected with siRNA targeting ZNF143 (siZNF143) or GFP (siCTRL). Control ChIP assays were performed with non-specific immunoglobulin G (IgG) and ZNF143 knockdown control with anti-ZNF143 antibodies on cells treated with siRNA targeting ZNF143. The gene symbols listed are those of the genes closest to the tested genomic region.
Finally, we determined the over-represented functional categories of genes located near loci occupied by both ZNF143 and Thap11 (36). As for the whole set of promoters recognized by ZNF143, the enriched terms concern cell growth and primary metabolism, whereas cell signalling and cell development were under-represented. The association of ZNF143 to the MLT1J retroelement via a SBS2 motif suggests a possible association of THAP11 to the same retroelement. Indeed, we verified by anti-HA ChIP-qPCR that THAP11 is also associated in vivo to an MLT1J retroelement locus (Supplementary Figure S12A).
Genome-wide analysis of Notch1/ICN1 binding in HPB-ALL cells reveals widespread overlap with ZNF143
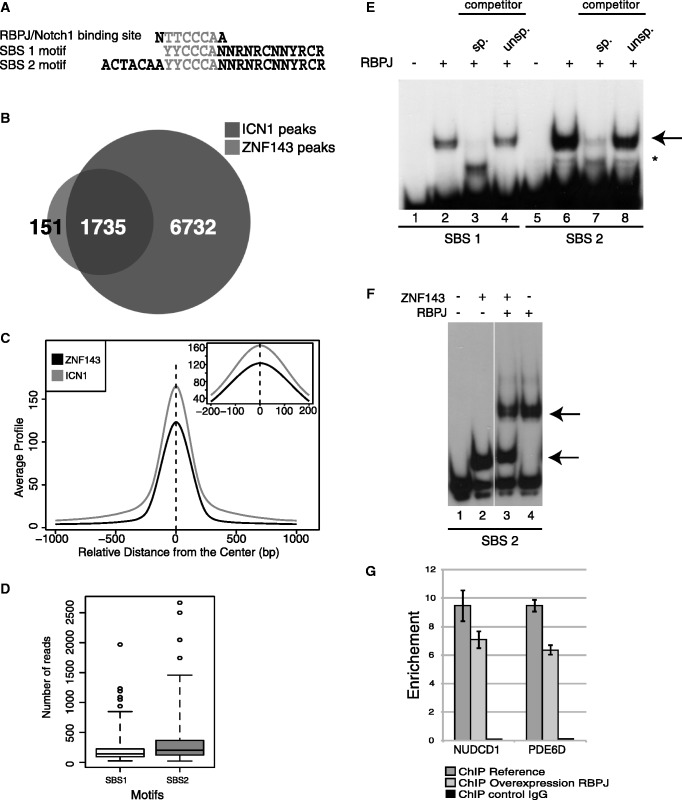
The ICN1 protein, the active form of the Notch1 receptor, is recruited to promoters by RBPJ, which binds to the TTCCCAA consensus motif (23) frequently associated to an ACTACAN motif (39). The striking identity of this extended motif with SBS2 suggests a possible overlap between the RBPJ/ICN1 and ZNF143-binding events (Figure 5A). To test this assumption, we performed a ChIP-Seq experiment in the HPB-ALL cell line using an antibody directed against ICN1 (Supplementary Table S1). The data were compared with the ZNF143-ChIP-Seq data from the same cells. We identified that among the 1886 ZNF143- and 8467 ICN1-binding events located in promoters (Supplementary Table S1), 91.9% of the ZNF143-binding events are shared with those of the ICN1 protein (Figure 5B and Supplementary Figure S13). On average, the positions of the ICN1 and ZNF143 co-peaks coincided exactly (Figure 5C), suggesting the recognition of common sites by the two proteins. We also evidenced, as for ZNF143, that ICN1-binding events have a higher enrichment signal when associated to an SBS2 than an SBS1 site (t-test: P < 10−5) (Figure 5D). By band shift assay, we showed that RBPJ binds specifically to SBS1 and SBS2 motifs, both containing the TTCCCA RBPJ-binding motif (Figure 5E). Surprisingly, we observed that the RBPJ-binding was more efficient on the SBS2 motif, suggesting the possible involvement of the ACTACAN motif in recognition. Band shift assays performed on SBS1 or SBS2 motifs in the presence of ZNF143 and RBPJ revealed that the binding of the two proteins is mutually exclusive (Figure 5F). This observation is also supported by the results of an in vivo competition assay showing that over-expression of RBPJ provides a significant decrease of the ZNF143 occupation on target promoters common to ZNF143 and RBPJ (Figure 5G). Globally, this suggests that the ZNF143-ICN1 co-peaks observed by ChIP-Seq did not result from a simultaneous co-occupation of the DNA by both proteins. Finally, the analysis of ICN1-ChIP-Seq data also revealed that the MLT1J subfamily of retroelements alone was significantly enriched in ICN1-binding events (Supplementary Figure S12B).
Figure 5.
ICN1-binding regions co-localize with the ZNF143-binding events. (A) Sequence alignment of the RBPJ-binding site, SBS1 and SBS2 motifs. (B) Venn diagram depicting the overlap of ZNF143 and ICN1-binding events in HPB-ALL cells. (C) Overlap of ICN1 sites with ZNF143-binding sites in HPB-ALL cells. Average profile of ICN1 and ZNF143 ChIP-Seq signals around the centre of all overlapping peaks. (D) Box blots showing ChIP-Seq score (number of reads) distribution of ICN1-binding events associated to SBS1 or SBS2 motifs. (E) RBPJ in vitro binding assays on SBS1 and SBS2 motifs. Gel retardation assays were performed with 32P labelled double stranded fragment containing the indicated motif. The labelled probe was incubated in the presence (+) or absence (−) of RBPJ. Reactions in lanes 3, 7 and 4, 8 were performed in the presence of an excess of unlabelled specific (sp.) or unspecific (unsp.) competitors. The RBPJ–DNA complex is indicated by an arrow, the asterisk denotes an unrelated product. (F) The binding of RBPJ and ZNF143 is mutually exclusive in vitro. Gel retardation assays were performed with 32P labelled double-stranded fragment containing the SBS2 motif in the presence (+) or absence (−) of the indicated proteins. Arrows indicate the RBPJ–DNA and ZNF143–DNA complexes. (G) The binding of RBPJ and ZNF143 is mutually exclusive in vivo. ZNF143-ChIP-qPCR enrichment analysis on promoters bound by ZNF143 and Notch1. ZNF143-ChIP was performed on non-induced (ChIP reference) and induced FLP_RBPJ-HA cells expressing the RBPJ-HA (ChIP over-expression RBPJ) protein. ChIP enrichment was measured by qPCR using specific primers on promoter compared with negative regions located at 2 kb. Primers are available on request. A control ChIP was performed using non-specific IgG antibodies (ChIP control IgG). The gene symbols listed are those of the genes closest to the tested genomic region.
ZNF143, THAP11 and ICN1 levels directly modulate expression of common target genes
The overlapping binding pattern of ZNF143, THAP11 and ICN1 led us to examine whether they play a direct role in the regulation of common target genes. To answer this question, we analysed the global or partial gene expression pattern after alteration of the level of the three factors. First, we performed global profiling of gene expression by RNA-Seq in cells depleted of or over-expressing ZNF143. Both the ZNF143 siRNA-mediated knockdown and over-expression efficiencies were affirmed at the protein (Figure 6A, lanes 1–6). After deep RNA sequencing, we examined the relative expression of the genes located at +/−2 kb from a ZNF143 peak (Supplementary Table S2). The analysis revealed two major populations of differentially expressed target genes. The largest population (837 genes), as expected, was downregulated after ZNF143 knockdown and broadly upregulated after ZNF143 over-expression (Figure 6B, left part). The second, smaller population (386 genes), was unexpectedly upregulated after ZNF143 knockdown and mostly downregulated following its over-expression (Figure 6B, right part). The general tendency of the profiles suggests that on the full set of genes, ZNF143 acts mainly as an activator but can also be involved in the repression of transcription. An independent verification of the RNA-Seq data by RT-qPCR on a set of 51 genes activated or repressed after ZNF143 knockdown, recapitulated the majority of the findings (Supplementary Figure S14A). The genes positively regulated by ZNF143 (downregulated after ZNF143 knockdown) are primarily associated with ontology terms involved in transcription (mainly zinc-finger DNA-binding proteins), cell cycle regulation and chromatin organization (Supplementary Figure S14B, red bars). On the other hand, the negatively regulated genes (upregulated after ZNF143 knockdown) were only slightly enriched in genes involved in transcription processes (Supplementary Figure S14B, blue bars).
Figure 6.
ZNF143, THAP11 and ICN1 co-regulate the expression of target genes. (A) Western blots showing (i) knockdown of ZNF143 at 72 h post-transfection of HeLa cells with specific siRNA (lane 3) and siRNA control (lanes 1 and 2); (ii) over-expression of ZNF143 in FLP143 cells at 12 h (lane 5), 24 h (lane 6) post-induction, uninduced cells (lane 4); (iii) knockdown of ICN1 at 48 h and 72 h post- treatment of HPB-ALL cells with 500 nM of GSI (compound E) (lanes 9 and 10), DMSO treated (lane 8) and non-treated cells (lane 7). Tubulin was used as a loading control. (B) Bar chart depicting the RNA-Seq expression profiling of all genes located at +/−2 kb of a ZNF143 peak summit, after knockdown or over-expression of ZNF143. In grey and black are represented the fold change of expression 48 h after siRNA-mediated knockdown and 24 h after induced over-expression of ZNF143 compared with control, respectively. The genes are ordered decreasingly according their positive or negative fold change expression after ZNF143 knockdown. (C) Bar chart depicting the relative gene expression ratio determined by RT-qPCR for genes containing SBS1 or SBS2 sites, after ICN1 and ZNF143 knockdown compared with control. RNA was extracted 72 h post-transfection of HeLa-S3 cells with specific siRNAs targeting ZNF143 and from HPB-ALL cells 72 h post-treatment with 500 nM of GSI (compound E). Values represent the mean +/− SD of two or three independent experiments normalized to the GAPDH level. The dark grey line corresponds to a 0.5-fold expression ratio (D) Bar chart depicting the relative gene expression ratio determined by RT-qPCR for genes containing SBS1 or SBS2 sites, after THAP11 and ZNF143 knockdown compared with control. RNA was extracted 72 h post-transfection of HeLa-S3 cells with specific siRNAs targeting ZNF143 or THAP11. Values represent the mean +/− SD of two or three independent experiments normalized to the GAPDH level. The dark grey line corresponds to a 0.5-fold expression ratio (E) Average ChIP-Seq profile of Pol II and (F) Average profile of H3K4me3 around the centre of ZNF143-binding events identified in HeLa cells at +/−2 kb from a TSS (Supplementary Table S4). All ZNF143-binding events, binding events containing only SBS1 or only SBS2, are indicated in black, dark grey and light grey, respectively. (G) RNA Pol II-ChIP-qPCR enrichment analysis on promoters bound by THAP11 and containing SBS2 sites. Pol II-ChIP was performed on cells treated with control siRNA (siCTRL), knockdown for ZNF143 (siZNF143) or knockdown for ZNF143 and over-expressing THAP11 (siZNF143 + THAP11-HA). ChIP enrichment was measured by qPCR using specific primers on promoter compared with negative regions located at 2 kb. Values represent the mean +/− SD of two or three independent experiments normalized to the GAPDH promoter levels. Primers are available on request. The gene symbols listed are those of the genes closest to the tested genomic region.
As we found THAP11 and ICN1 associated with ZNF143-binding motifs, we next analysed by RT-qPCR the effect of THAP11 and ICN1 knockdown on 51 ZNF143 target genes. After THAP11 knockdown in HeLa-S3 cells using specific siRNA (Figure 6D), 28 of the 51 genes were significantly upregulated and 4 of the 51 downregulated, in agreement with the major repressor function of the factor. After knockdown of ICN1 (Figure 6A, lanes 7–10), by γ-secretase inhibitor (GSI) treatment on HPB-ALL cells, 6 of the 51 genes were significantly upregulated and 20 of the 51 downregulated (Figure 6C). Combining our expression data on this subset of genes, we observed that both SBS1- or SBS2-containing genes are generally positively regulated by ZNF143 and ICN1, but, in contrast, genes carrying only SBS2 sites are significantly upregulated by THAP11 knockdown (Figure 6D). We have noticed the presence of genes upregulated by both ZNF143 and THAP11 knockdown. This is exemplified by JUN that possesses an SBS1 site and ALG14 with an SBS2 site in their promoters (Figure 6D). This upregulation suggests a direct or indirect negative regulation effect mediated by both factors. Taken together, our results identify a set of genes that can be differentially regulated by loss or gain of ZNF143, THAP11 and ICN1 function.
Promoters with associated SBS1 or SBS2 motifs carry different levels of RNA polymerase II and histone activation marks
The aforementioned data revealed that SBS1- and SBS2-mediated modulation of gene expression involves a competitive binding of different transcription factors on these motifs. We sought then to understand whether this differential occupation could be correlated with variations in Pol II occupancy and various histone H3 methylation and acetylation marks associated to activation or repression of transcription. It is well established that in the promoter of Pol II transcribed genes, the activating H3K4me3 and H3K27ac marks form two peaks on either side of the TSS, whereas the H3K36me3 mark accumulates within the transcribed region (40). Enhancers are commonly characterized by the presence of both H3K27ac and H3K4me1 histone marks (41). In contrast, the repressive marks H3K9me3 and H3K27me3 are associated to the inactive state of the chromatin (40). We first examined in HeLa-S3 cells the presence of H3K27ac and H3K4me1 around all ZNF143 peaks: ZNF143 peaks in promoters (+/−2 kb of a TSS) and ZNF143 peaks outside of promoters (Supplementary Figure S15). ZNF143 enriched regions outside of promoters do not co-localize with H3K27ac and H3K4me1 enhancer marks (Supplementary Figure S15C). On the contrary, peaks located in promoters are all highly enriched in ZNF143 and H3K27ac activation marks (Supplementary Figure S15B). Subsequently, we examined in HeLa-S3 cells the relation between histone H3 trimethylation and Pol II occupancy patterns on the ZNF143-binding events located near TSS. Heat maps representing the enrichment reveal a broad co-localization of ZNF143, Pol II and H3K4me3 modifications (Supplementary Figure S16A). Similarly, the H3K36me3 modification is found on the body of the genes associated to ZNF143-binding events (Supplementary Figure S16B). In contrast to H3K4me3, there was no detectable H3K9me3 and H3K27me3 modification associated to ZNF143-binding events (Supplementary Figure S16A). We observed a differential behaviour of loci associated to SBS1 or SBS2 motifs with respect to Pol II, H3K4me3 and H3K36me3 enrichments. Indeed, in opposition to ZNF143 enrichment (Figure 2D and Supplementary Figure S16C), the Pol II (Figure 6E), H3K4me3 (Figure 6F) and H3K36me3 enrichments (Supplementary Figure S16B) are higher on SBS1 than on SBS2-associated loci (t-test: P < 10−5). Furthermore, the Pol II occupancy profile forms two peaks on each side of SBS2 loci, whereas a single one is observed for loci associated to SBS1 (Figure 6E). A similar behaviour of Pol II and H3K4me3 on SBS1 and SBS2 sites is observed for two different ChIP-Seq data sets from K562 and mES cells (Supplementary Figure S16D). The aforementioned observations suggest a distinct transcriptional control on SBS2 sites owing to the preferential binding of THAP11 on the SBS2 motif. We tested by ChIP-qPCR the Pol II occupation on several promoters containing SBS2 motifs after ZNF143 siRNA-mediated knockdown associated or not to THAP11 overproduction (Figure 6G). As expected, the results show a decrease of Pol II occupancy after ZNF143 knockdown, and the Pol II deficiency is amplified after THAP11 overexpression (Figure 6G).
DISCUSSION
In this study, we obtained a set of >3000 genes that may be subject to a ZNF143-mediated regulation in both humans and mouse. These genes are globally highly enriched in terms related to transcription, cell cycle, RNA processing and translation processes, whereas cell signalling and cell development were under-represented. In addition, we observed as reported for PC3 cells (37) that the ZNF143 knockdown reduces significantly the cell proliferation rate in HeLa cells. These observations strongly suggest a particular role for this factor in cell growth regulation. We also report that both activation and repression of gene expression directed by ZNF143 occur in human cells.
Among the ZNF143 target genes, it is worth mentioning 114 KRAB zinc-finger transcription factors located in tandem on human chromosome 19. Considering the amino acid sequence similarity between the subdomain B of the KRAB box and the ncRNA transcription activation motif of ZNF143 (42), it is tempting to speculate that ZNF143 could originate from an ancient KRAB family member. A co-evolutionary link exists between retroelements and the presence of tandem zinc-finger genes (43). If we take into consideration the SBS2 site embedded in MLT1J retrotransposons, we can hypothesize that ZNF143 have diverged into a transcriptional regulator of more recent genes of the same family, generated by the successive invasion waves of genomes by KRAB zinc-fingers (44).
According to our results, ZNF76, the vertebrate paralog of ZNF143, occupies exactly the same binding sites as those recognized by ZNF143. We previously showed that ZNF76 is able to activate transcription (16), but other results have also identified ZNF76 as a transcriptional repressor (38). Having the same DNA-binding specificity as ZNF143, ZNF76 could have a different non-redundant function or it could play an auxiliary replacement function in particular situations.
As many ZNF143-binding events are located at proximity of ncRNA genes, this raises the question of the functionality of these events in ncRNA gene transcription. For the ZNF143 occupancy on 12 Pol III type 3 and 44 Pol II snRNA genes, it is very likely that the factor is involved in their transcriptional activation. Indeed, as very recently reported (45), all of these promoters contain the canonical sequence elements (35) and are bound by their proteins counterparts (46). Thirteen miRNA genes and a large number of snoRNA and scaRNA genes, found near ZNF143-binding events, are located in the first exon or in the first intron of coding or non-coding host genes. This opens the possibility that ZNF143 is involved in the expression of these ncRNAs by directing expression of the host gene. This is observed for MIR632, MIR659, MIR663, MIR933, MIR1292 and MIR3917 for which the host genes are deregulated in cells depleted or over-expressing ZNF143.
A major aspect of our study concerns the identification and the functional characterization of the SBS1 (18 bp) and SBS2 (25 bp) motifs associated to ZNF143-binding events. The additional ACTACAN sequence found in SBS2 has already been reported as part of one of the most represented motifs—the M4 motif—in gene promoters (47) and is not required for ZNF143 binding. It comes out from our study that the TBS ACTACAATTCCCAG is always physically linked to an overlapping ZNF143-binding site at the whole genome scale. We also evidenced, in vitro as well as in vivo, that the binding of ZNF143 and THAP11 on the SBS2 motif is mutually exclusive. These observations clearly indicate that the SBS2 subset of ZNF143-binding sites contains the set of TBSs. Moreover, we also demonstrated that the two proteins are both involved in the transcriptional regulation of the same set of genes. This gene expression modulation is achieved through a mutually exclusive binding on overlapping binding sites. As the result of the competitive binding of the two factors and to the main repressor activity of THAP11 (25), the promoters containing SBS2 sites have lower Pol II and H3K4/36me3 ChIP-Seq profiles. Nevertheless, unlike Pol II and H3K4me3 marks, ZNF143 shows a notably higher ChIP-Seq enrichment on SBS2 sites than on SBS1 sites. This observation supports the notion that SBS1- and SBS2-containing promoters are in different regulatory contexts.
The examination of SBS1 and SBS2 sites led us to consider another overlapping binding event with ICN1. Indeed, we demonstrated that in the T-lymphoblastic leukaemia cell line HPB-ALL, the constitutively over-expressed ICN1 protein is recruited in 27.6% of the ICN1-binding events via the TTCCCA sequence embedded in SBS1 and SBS2 motifs. In addition, we demonstrated that the RBPJ protein, which recruits ICN1, binds in vitro as well as in vivo to the SBS1 and SBS2 sites in an exclusive manner in the presence of ZNF143. Another recent study performed in a different T-lymphoblastic leukemia cell line (CUTLL1) also highlighted that the ICN1-binding profile is partially overlapping ZNF143-binding sites (48). However, the authors of this study identified only an SBS2-type site as being bound by ZNF143. In contrast, our work performed on seven different human and mouse cell types clearly distinguishes the two motifs SBS1 and SBS2 as bound by ZNF143 and subjected to differential binding by two other unrelated transcription factors. As for ZNF143, our ChIP-Seq data on ICN1 showed higher tag enrichment for peaks associated to the SBS2 than to the SBS1 sites. This is consistent with our hypothesis that promoters containing either the SBS1 or SBS2 site are differently regulated. The gene expression analysis after GSI treatment in HPB-ALL cells confirmed the major activation function of ICN1 binding on both SBS1 and SBS2 sites.
We can infer from our results that ZNF143, THAP11 and ICN1/RBPJ are not only involved in a competitive binding on overlapping sites but are also involved in the transcriptional activity of adjacent genes. We revealed in this study that the genes targeted by ZNF143 are mainly involved in biological processes related to cell growth such as transcription, translation and cell cycle. Our observation is consistent with several reports concerning the role of THAP11 and ICN1. Indeed, the capacity of THAP11 to exert anti-differentiation effects in mES cells was originally suggested from THAP11 global dependent transcriptional repression of multiple genes that are either directly or indirectly involved in differentiation (27). In contrast, more recent results in mES cells showed that THAP11 can either activate or repress expression of its transcriptional target genes, essentially involved in the growth of embryonic stem cells (26). ICN1 is constitutively over-expressed in acute T-cell lymphoblastic leukaemia cells where it deregulates genes involved in cell growth processes (48,49). ZNF143, as shown in this study, is also involved in the regulation of genes involved in cell growth and has been shown to be important to maintain the undifferentiated state of mES cells (19). Taken together, it appears that ZNF143, THAP11 and ICN1 could together be important for the transcriptional modulation of genes involved in the proliferation of rapidly dividing cells, like cancer cells and embryonic stem cells. Therefore, this control is exerted via a competitive binding on overlapping binding sites.
All the observations and results taken together led us to propose the following model (Figure 7). The competitive and combinational binding of ZNF143, THAP11 and RBPJ/ICN1 on single or multiple SBS1 and SBS2 sites in promoters is directly involved in the transcriptional regulation of target genes. This is achieved by activation via ZNF143 and repression via THAP11 and RBPJ. Moreover, in HPB-ALL cells, the over-expressed ICN1 recruited by the repressor RBPJ competes for activation either with ZNF143 on SBS1 motifs or with THAP11 and ZNF143 on SBS2 motifs (Figure 7). Nevertheless, our model is restricted to certain genes being activated by ZNF143 and repressed by THAP11. Other genes, such as ALG14 and JUN, are negatively regulated by both THAP11 and ZNF143. This suggests an additional modulation mechanism that involves probably other partners acting positively or negatively on transcription. Such an additional partner could be ZNF76 that binds exactly the same motif as ZNF143. This extra layer of complexity should be further investigated.
Figure 7.
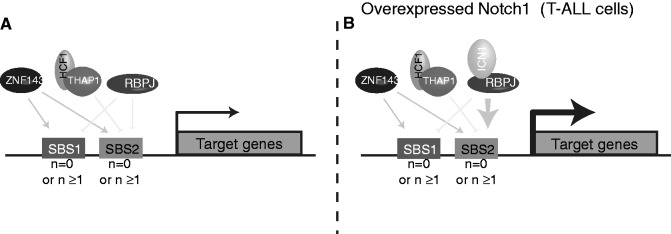
A model for modulation of gene expression by ZNF143, THAP11 and ICN1. The model considers single or multiple SBS1 and/or SBS2 motifs (n = 0 or n ≥ 1) in promoters. ZNF143 activates the expression of genes through both SBS1 and SBS2 sites. THAP11 represses the expression of genes only through SBS2 sites. Instead, genes that are regularly repressed by RBPJ through SBS1 and SBS2 sites (A) are activated in T-ALL cells when ICN1 is over-expressed (B).
Another interesting outcome of this work concerns the evolutionary history of SBS2 sites. As shown for several transcription factors (50), it emerged from our data that the very ancient family of MLT1J retro-transposons could be at the origin of the SBS2 sites. Furthermore, a correlation exists between KRAB zinc-finger gene expansions and retro-transposition waves (43). We thus speculate that ZNF143 originated from an ancient KRAB zinc-finger transcription factor family targeting at the time still active MLT1J retro-transposons. In the course of evolution, these events could have caused expansion of the repertoire of ZNF143, THAP11 and ICN1 regulated genes by spreading out the SBS2 sites in promoter regions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–7, Supplementary Figures 1–16 and Supplementary Materials and Methods.
FUNDING
Université de Strasbourg, the Centre National de la Recherche Scientifique, the Ministère de l'Enseignement Supérieur et de la Recherche (to R.P.N.M.), the Association pour la Recherche contre le Cancer (to R.P.N.M.) and the Ligue Contre le Cancer (CCIR-GE). Funding for open access charge: The Ligue Contre le Cancer (CCIR-GE).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful to Prof. B. Kempkes for the gift of the RBPJ construct, Dr. V. Penard-Lacronique for supplying the HPB-ALL cells, Dr. P. Assmann for mouse fibroblasts, Dr. I Davidson and S. Urban for the mouse embryonic stem cells, Dr. S. Pffefer for supplied material and Dr. A. Krol for critical reading of the manuscript. They also thank B. Jost, S. Le Gras and C. Keime from the sequencing platform of IGBMC (Illkirch, France); S. Baudrey and A. Schweigert for valuable technical assistance. They are thankful for the contribution of Y.N. Anno, O. Lecompte and O. Poch to the initial phase of the project. They are also grateful to E. Westhof for his interest in their study.
The overall study was conceived, designed and coordinated by P.C., with important contributions from R.P.N.-M. and E.M. R.P.N.-M. and E.M. performed the experiments. R.P.N.-M. analysed the sequencing data. J.C.A. contributed new reagents. R.P.N.-M. designed the figures. P.C. and R.P.N.-M. wrote the article with helpful contribution from E.M. and J.C.A.
REFERENCES
- 1.Vaquerizas JM, Kummerfeld SK, Teichmann SA, Luscombe NM. A census of human transcription factors: function, expression and evolution. Nat. Rev. Genet. 2009;10:252–263. doi: 10.1038/nrg2538. [DOI] [PubMed] [Google Scholar]
- 2.Lenhard B, Sandelin A, Carninci P. Regulatory elements: Metazoan promoters: emerging characteristics and insights into transcriptional regulation. Nat. Rev. Genet. 2012;13:233–245. doi: 10.1038/nrg3163. [DOI] [PubMed] [Google Scholar]
- 3.Spitz F, Furlong EEM. Transcription factors: from enhancer binding to developmental control. Nat. Rev. Genet. 2012;13:613–626. doi: 10.1038/nrg3207. [DOI] [PubMed] [Google Scholar]
- 4.Macquarrie KL, Fong AP, Morse RH, Tapscott SJ. Genome-wide transcription factor binding: beyond direct target regulation. Trends Genet. 2011;27:141–148. doi: 10.1016/j.tig.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rosenbloom KR, Dreszer TR, Long JC, Malladi VS, Sloan CA, Raney BJ, Cline MS, Karolchik D, Barber GP, Clawson H, et al. ENCODE whole-genome data in the UCSC Genome Browser: update 2012. Nucleic Acids Res. 2012;40:D912–D917. doi: 10.1093/nar/gkr1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Consortium,T.E.P., Consortium,T.E.P., data analysis coordination,O.C., data production,D.P.L., data analysis,L.A., group,W., scientific management,N.P.M., steering committee,P.I., Boise State University and University of North Carolina at Chapel Hill Proteomics groups (data production and analysis), Broad Institute Group (data production and analysis) et al. (2012) An integrated encyclopedia of DNA elements in the human genome. Nature, 488, 57–74.
- 7.Walhout AJM. Unraveling transcription regulatory networks by protein-DNA and protein-protein interaction mapping. Genome Res. 2006;16:1445–1454. doi: 10.1101/gr.5321506. [DOI] [PubMed] [Google Scholar]
- 8.Farnham PJ. Insights from genomic profiling of transcription factors. Nat. Rev. Genet. 2009;10:605–616. doi: 10.1038/nrg2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wilson NK, Foster SD, Wang X, Knezevic K, Schütte J, Kaimakis P, Chilarska PM, Kinston S, Ouwehand WH, Dzierzak E, et al. Combinatorial transcriptional control in blood stem/progenitor cells: genome-wide analysis of ten major transcriptional regulators. Cell Stem Cell. 2010;7:532–544. doi: 10.1016/j.stem.2010.07.016. [DOI] [PubMed] [Google Scholar]
- 10.Schaub M, Myslinski E, Schuster C, Krol A, Carbon P. Staf, a promiscuous activator for enhanced transcription by RNA polymerases II and III. EMBO J. 1997;16:173–181. doi: 10.1093/emboj/16.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Myslinski E, Amé JC, Krol A, Carbon P. An unusually compact external promoter for RNA polymerase III transcription of the human H1RNA gene. Nucleic Acids Res. 2001;29:2502–2509. doi: 10.1093/nar/29.12.2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gérard M-A, Myslinski E, Chylak N, Baudrey S, Krol A, Carbon P. The scaRNA2 is produced by an independent transcription unit and its processing is directed by the encoding region. Nucleic Acids Res. 2010;38:370–381. doi: 10.1093/nar/gkp988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hernandez-Negrete I, Sala-Newby GB, Perl A, Kunkel GR, Newby AC, Bond M. Adhesion-dependent Skp2 transcription requires selenocysteine tRNA gene transcription-activating factor (STAF) Biochem. J. 2011;436:133–143. doi: 10.1042/BJ20101798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gérard M-A, Krol A, Carbon P. Transcription factor hStaf/ZNF143 is required for expression of the human TFAM gene. Gene. 2007;401:145–153. doi: 10.1016/j.gene.2007.07.011. [DOI] [PubMed] [Google Scholar]
- 15.Myslinski E, Gérard M-A, Krol A, Carbon P. Transcription of the human cell cycle regulated BUB1B gene requires hStaf/ZNF143. Nucleic Acids Res. 2007;35:3453–3464. doi: 10.1093/nar/gkm239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Myslinski E, Krol A, Carbon P. ZNF76 and ZNF143 are two human homologs of the transcriptional activator Staf. J. Biol. Chem. 1998;273:21998–22006. doi: 10.1074/jbc.273.34.21998. [DOI] [PubMed] [Google Scholar]
- 17.Wakasugi T, Izumi H, Uchiumi T, Suzuki H, Arao T, Nishio K, Kohno K. ZNF143 interacts with p73 and is involved in cisplatin resistance through the transcriptional regulation of DNA repair genes. Oncogene. 2007;26:5194–5203. doi: 10.1038/sj.onc.1210326. [DOI] [PubMed] [Google Scholar]
- 18.Halbig KM, Lekven AC, Kunkel GR. The transcriptional activator ZNF143 is essential for normal development in zebrafish. BMC Mol Biol. 2012;13:3. doi: 10.1186/1471-2199-13-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen X, Fang F, Liou Y-C, Ng H-H. Zfp143 regulates Nanog through modulation of Oct4 binding. Stem Cells. 2008;26:2759–2767. doi: 10.1634/stemcells.2008-0398. [DOI] [PubMed] [Google Scholar]
- 20.Chia N-Y, Chan Y-S, Feng B, Lu X, Orlov YL, Moreau D, Kumar P, Yang L, Jiang J, Lau M-S, et al. A genome-wide RNAi screen reveals determinants of human embryonic stem cell identity. Nature. 2010;468:316–320. doi: 10.1038/nature09531. [DOI] [PubMed] [Google Scholar]
- 21.Kopan R, Ilagan MXG. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ranganathan P, Weaver KL, Capobianco AJ. Notch signalling in solid tumours: a little bit of everything but not all the time. Nat. Rev. Cancer. 2011;11:338–351. doi: 10.1038/nrc3035. [DOI] [PubMed] [Google Scholar]
- 23.Grabher C, Boehmer, von H, Look AT. Notch 1 activation in the molecular pathogenesis of T-cell acute lymphoblastic leukaemia. Nat. Rev. Cancer. 2006;6:347–359. doi: 10.1038/nrc1880. [DOI] [PubMed] [Google Scholar]
- 24.Bray SJ. Notch signalling: a simple pathway becomes complex. Nat. Rev. Mol. Cell Biol. 2006;7:678–689. doi: 10.1038/nrm2009. [DOI] [PubMed] [Google Scholar]
- 25.Dejosez M, Levine SS, Frampton GM, Whyte WA, Stratton SA, Barton MC, Gunaratne PH, Young RA, Zwaka TP. Ronin/Hcf-1 binds to a hyperconserved enhancer element and regulates genes involved in the growth of embryonic stem cells. Genes Dev. 2010;24:1479–1484. doi: 10.1101/gad.1935210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parker JB, Palchaudhuri S, Yin H, Wei J, Chakravarti D. A Transcriptional Regulatory Role of the THAP11-HCF-1 Complex in Colon Cancer Cell Function. Mol. Cell. Biol. 2012;32:1654–1670. doi: 10.1128/MCB.06033-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dejosez M, Krumenacker JS, Zitur LJ, Passeri M, Chu L-F, Songyang Z, Thomson JA, Zwaka TP. Ronin is essential for embryogenesis and the pluripotency of mouse embryonic stem cells. Cell. 2008;133:1162–1174. doi: 10.1016/j.cell.2008.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anno Y-N, Myslinski E, Ngondo-Mbongo RP, Krol A, Poch O, Lecompte O, Carbon P. Genome-wide evidence for an essential role of the human Staf/ZNF143 transcription factor in bidirectional transcription. Nucleic Acids Res. 2011;39:3116–3127. doi: 10.1093/nar/gkq1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maier S, Santak M, Mantik A, Grabusic K, Kremmer E, Hammerschmidt W, Kempkes B. A somatic knockout of CBF1 in a human B-cell line reveals that induction of CD21 and CCR7 by EBNA-2 is strictly CBF1 dependent and that downregulation of immunoglobulin M is partially CBF1 independent. J. Virol. 2005;79:8784–8792. doi: 10.1128/JVI.79.14.8784-8792.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics (Oxford, England) 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu T, Ortiz JA, Taing L, Meyer CA, Lee B, Zhang Y, Shin H, Wong SS, Ma J, Lei Y, et al. Cistrome: an integrative platform for transcriptional regulation studies. Genome Biol. 2011;12:R83. doi: 10.1186/gb-2011-12-8-r83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hernandez N. Small nuclear RNA genes: a model system to study fundamental mechanisms of transcription. J. Biol. Chem. 2001;276:26733–26736. doi: 10.1074/jbc.R100032200. [DOI] [PubMed] [Google Scholar]
- 36.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 37.Izumi H, Wakasugi T, Shimajiri S, Tanimoto A, Sasaguri Y, Kashiwagi E, Yasuniwa Y, Akiyama M, Han B, Wu Y, et al. Role of ZNF143 in tumor growth through transcriptional regulation of DNA replication and cell-cycle-associated genes. Cancer Sci. 2010;101:2538–2545. doi: 10.1111/j.1349-7006.2010.01725.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zheng G, Yang Y-C. ZNF76, a novel transcriptional repressor targeting TATA-binding protein, is modulated by sumoylation. J. Biol. Chem. 2004;279:42410–42421. doi: 10.1074/jbc.M407287200. [DOI] [PubMed] [Google Scholar]
- 39.Ettwiller L, Paten B, Ramialison M, Birney E, Wittbrodt J. Trawler: de novo regulatory motif discovery pipeline for chromatin immunoprecipitation. Nat. Methods. 2007;4:563–565. doi: 10.1038/nmeth1061. [DOI] [PubMed] [Google Scholar]
- 40.Barski A, Cuddapah S, Cui K, Roh T-Y, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 41.Maston GA, Landt SG, Snyder M, Green MR. Characterization of enhancer function from genome-wide analyses. Annu. Rev. Genom. Hum. Genet. 2012;13:29–57. doi: 10.1146/annurev-genom-090711-163723. [DOI] [PubMed] [Google Scholar]
- 42.Schuster C, Krol A, Carbon P. Two distinct domains in Staf to selectively activate small nuclear RNA-type and mRNA promoters. Mol. Cell Biol. 1998;18:2650–2658. doi: 10.1128/mcb.18.5.2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thomas JH, Schneider S. Coevolution of retroelements and tandem zinc finger genes. Genome Res. 2011;21:1800–1812. doi: 10.1101/gr.121749.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Emerson RO, Thomas JH. Adaptive evolution in zinc finger transcription factors. PLoS Genet. 2009;5:e1000325. doi: 10.1371/journal.pgen.1000325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.James Faresse N, Canella D, Praz V, Michaud J, Romascano D, Hernandez N. Genomic study of RNA polymerase II and III SNAPc-bound promoters reveals a gene transcribed by both enzymes and a broad use of common activators. PLoS Genet. 2012;8:e1003028. doi: 10.1371/journal.pgen.1003028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.White RJ. Transcription by RNA polymerase III: more complex than we thought. Nat. Rev. Genet. 2011;12:459–463. doi: 10.1038/nrg3001. [DOI] [PubMed] [Google Scholar]
- 47.Xie X, Lu J, Kulbokas EJ, Golub TR, Mootha V, Lindblad-Toh K, Lander ES, Kellis M. Systematic discovery of regulatory motifs in human promoters and 3' UTRs by comparison of several mammals. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang H, Zou J, Zhao B, Johannsen E, Ashworth T, Wong H, Pear WS, Schug J, Blacklow SC, Arnett KL, et al. Genome-wide analysis reveals conserved and divergent features of Notch1/RBPJ binding in human and murine T-lymphoblastic leukemia cells. Proc. Natl Acad. Sci. USA. 2011;108:14908–14913. doi: 10.1073/pnas.1109023108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Palomero T, Lim WK, Odom DT, Sulis ML, Real PJ, Margolin A, Barnes KC, O'Neil J, Neuberg D, Weng AP, et al. NOTCH1 directly regulates c-MYC and activates a feed-forward-loop transcriptional network promoting leukemic cell growth. Proc. Natl Acad. Sci. USA. 2006;103:18261–18266. doi: 10.1073/pnas.0606108103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jordan IK, Rogozin IB, Glazko GV, Koonin EV. Origin of a substantial fraction of human regulatory sequences from transposable elements. Trends Genet. 2003;19:68–72. doi: 10.1016/s0168-9525(02)00006-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.