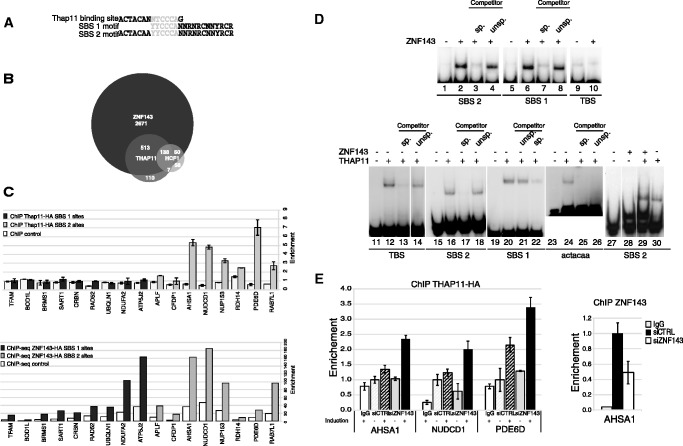
Figure 4.
Overlapping of ZNF143 and Thap11 DNA-binding events. (A) Sequence alignment of the TBS, SBS1 and SBS2 motifs. The sequence indicated in light grey is shared by the three motifs. (B) Venn diagram depicting the overlap of ZNF143/Zfp143-, Thap11- and Hcf1-binding events in mouse ES cells. The analysis was performed using the complete data set listed in Supplementary Table S5 and published ChIP-Seq data (25). (C) Histograms depicting the enrichment of genomic regions containing only SBS1 or SBS2 motifs in ChIP samples. ChIP assays with anti-HA antibody were performed on induced FLP143-HA and FLPTHAP11-HA stable cell lines. Uninduced stable cell lines were used as control. THAP11-HA data are obtained from ChIP-qPCR experiment, amplifying a promoter region containing the binding site. Primer sequences are available on request. ZNF143-HA data are obtained from ChIP-Seq experiment, using read number values in regions amplified by qPCR for THAP11-HA, expanded to 200 bp. SBS1 sites and SBS2 sites correspond to promoters containing only the SBS1 motif or only the SBS2 motif, respectively. Fold enrichment in promoters was normalized to control regions located at 2 kb of tested promoters. The gene symbols listed are those of the genes closest to the tested genomic region. (D) ZNF143 and THAP11 in vitro binding assays on SBS1, SBS2, TBS and TBS sub-motif. Gel retardation assays were performed with 32P labelled double-stranded fragment containing the indicated motif. The labelled probe was incubated in the presence (+) or absence (−) of ZNF143 or THAP11 DBD. The reactions in lanes 3, 7, 13, 17, 22, 25 and 4, 8, 14, 18, 21 were performed in the presence of an excess of unlabelled specific (sp.) or unspecific (unsp.) competitors. The ACTACAA probe corresponds to the 5′part of the TBS. Each lane presented in a single panel of the gel picture was from the same gel and the same exposure of the autoradiogram. (E) Histograms depicting the enrichment of promoter regions containing SBS2 motifs in ChIP samples. ChIP assays with anti-HA antibody were performed on induced or non-induced FLPTHAP11-HA stable cell line, transfected with siRNA targeting ZNF143 (siZNF143) or GFP (siCTRL). Control ChIP assays were performed with non-specific immunoglobulin G (IgG) and ZNF143 knockdown control with anti-ZNF143 antibodies on cells treated with siRNA targeting ZNF143. The gene symbols listed are those of the genes closest to the tested genomic region.