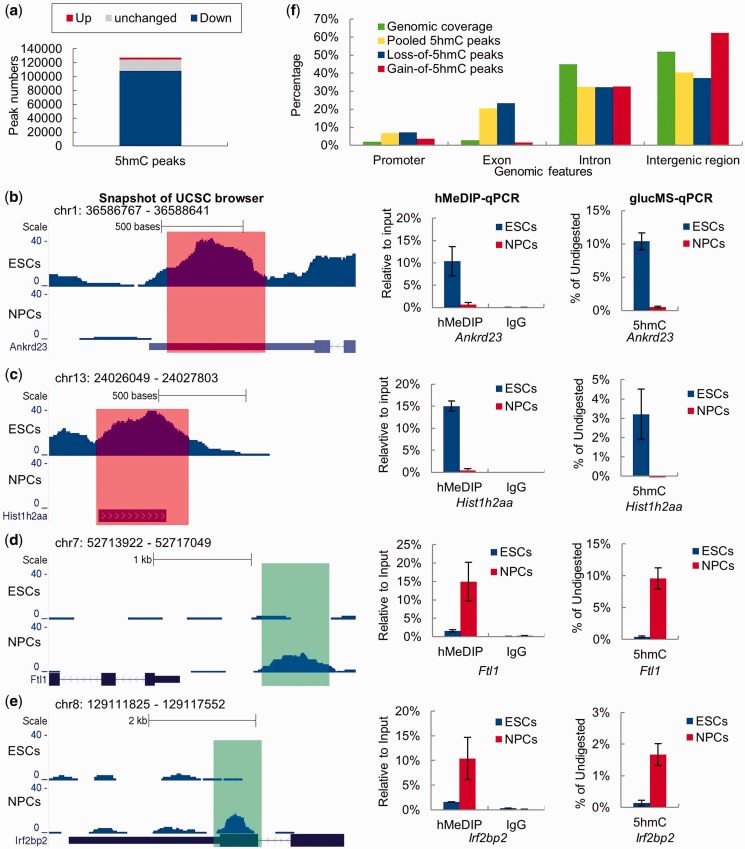
Figure 4.
Identification of DHMRs between ESCs and NPCs. (a) All 5hmC peaks called from the pooled hMeDIP-seq data were separated into three groups: down-regulated (log2 5hmC density ratio ≤ −1), up-regulated (log2 5hmC density ratio ≥ 1) and no significant change (−1 < log2 5hmC density ratio < 1) 5hmC peaks. (b–e) UCSC genome (mmp9) browser screen shots, hMeDIP-qPCR and glucMS-qPCR validation (mean values ± SD, n = 3) of two representative DHMRs with de-hydroxymethylation (Ankrd23 and Hist1h2aa) and two representative DHMRs with de novo hydroxymethylation (Ftl1 and Irf2bp2). (f) Percentages of pooled 5hmC peaks, ‘gain of 5hmC’ peaks and ‘loss of 5hmC’ peaks located at different genomic features.