Table 1.
Speedup due to targeted alignment
| NOTCH1 |
Cancer genes |
|||
|---|---|---|---|---|
| Sample | 1 core | 4 cores | 1 core | 4 cores |
| RNA-seq (2 × 50 bp) | 96 | 53 | 6.8 | 6.7 |
| RNA-seq (1 × 75 bp) | 92 | 25 | 4.5 | 4.0 |
| Exome-seq (2 × 100 bp) | 43 | 16 | 3.0 | 3.1 |
| WG-seq (2 × 100 bp) | 66 | 19 | 2.8 | 2.6 |
Speedups computed by comparing the time required to perform the full alignment and the combined time of triage classification plus mapping of the selected reads. For RNA-seq samples, target regions consisted of exons and 20 bp flanking sequences, s = 14 and H = 46. For exome and WG samples, which have longer reads, target regions consisted of exons and 85 bp flanking regions, s = 14 and H = 85. (Speedups are approximate with 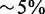 error; results may also vary depending on options used, background load, etc.)
error; results may also vary depending on options used, background load, etc.)
