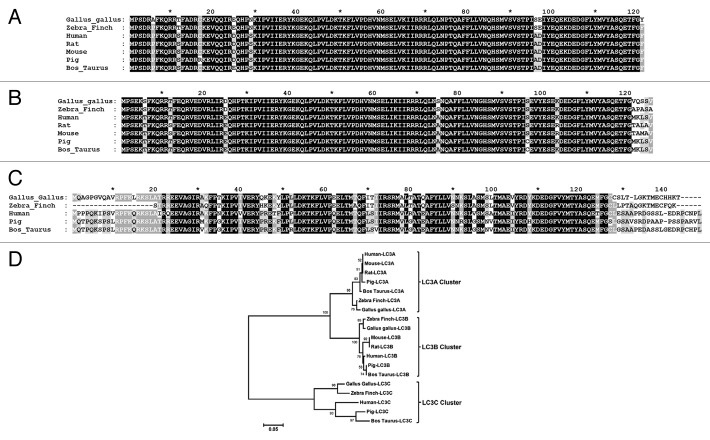
Figure 1. Comparison of the LC3A, LC3B and LC3C protein amino acid sequences derived from different species. The individual amino acid sequences of the (A) LC3A, (B) LC3B and (C) LC3C proteins were identified in the NCBI database, aligned using ClustalX version 2.135 and compared using GeneDoc Multiple Sequence Alignment Editor and Shading Utility version 2.7.001 (www.nrbsc.org/gfx/genedoc/). The LC3A protein accession numbers used were Gallus gallus XP_417327.2, Zebra Finch NP_001232518.1, Human NP_115903.1, Mouse NP_080011.1, Rat NP_955794.1, Pig NP_001164298.1, Bos taurus NP_001039640.1. The LC3B protein accession numbers used were Gallus gallus NP_001026632.1, Zebra Finch NP_001232222.1, Human NP_073729.1, Mouse NP_080436.1, Rat NP_074058.2, Pig NP_001177219.1, Bos taurus NP_001001169.1. The LC3C protein accession numbers used were Gallus gallus XP_419549.2, Human NP_001094528.1, Pig XP_003130606.1, Bos taurus NP_001094528.1 and Zebra Finch database ID TR:H0ZK62_TAEGU. (D) Phylogenetic analysis of the three LC3 paralogs following clustalX alignment of all the LC3 protein sequences. Phylogenetic analysis was conducted using MEGA4 version 436 in which the clustalX alignment was analyzed by the neighbor joining method using a bootstrap of 500 replications and a random seed of 64238.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.