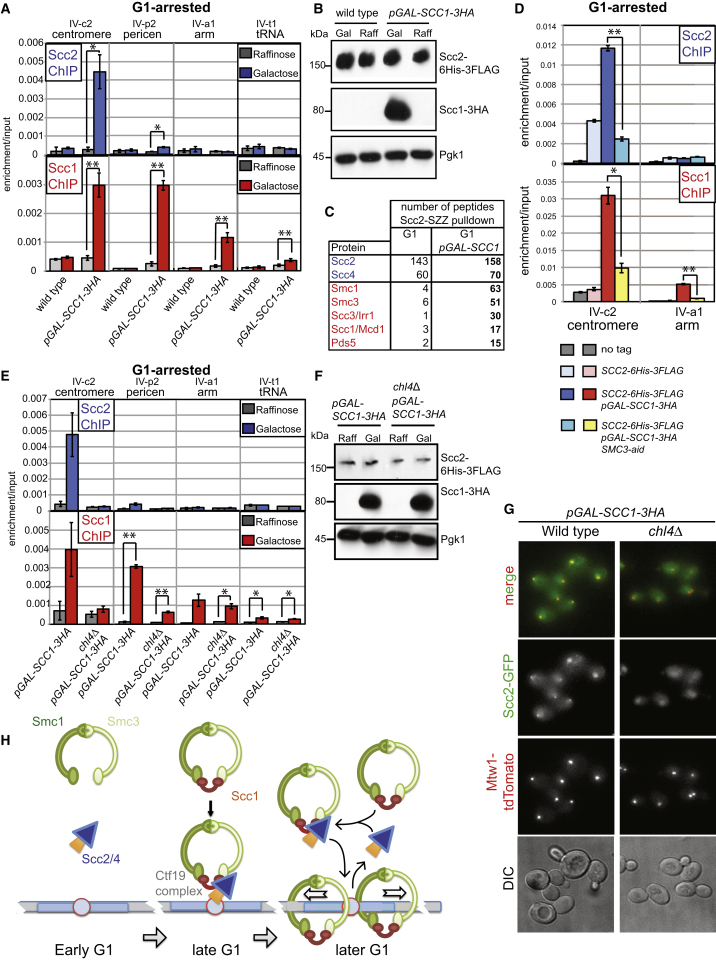
Figure 4.
SCC1 Expression Is Sufficient for Scc2 to Associate with Centromeres and for Scc1 Loading in G1
(A and B) Strains AM9335 (SCC2-6HIS-3FLAG) and AM9334 (SCC2-6HIS-3FLAG pGAL-SCC1-3HA) were arrested in G1 in raffinose-containing medium with alpha factor for 3 hr. Samples were extracted for anti-FLAG, anti-HA, and anti-Pgk1 immunoblotting (B) and ChIP-qPCR (A; raffinose), then 2% galactose was added to the remainder, while maintaining the G1 arrest. After 45 min the remainder of the culture was harvested for immunoblotting (B) and ChIP-qPCR (A; galactose).
(C) Mass spectrometry analysis of proteins copurifying with Scc2 after immunoprecipitation from G1-arrested cells with and without SCC1 expression. The number of identified peptides for the indicated proteins is shown.
(D) Smc3 is required for Scc2 and Scc1 to associate with centromeres. Strains AM1176 (no tag), AM6006 (SCC2-6HIS-3FLAG), AM9334 (SCC2-6HIS-3FLAG pGAL-SCC1), and AM11092 (SCC2-6HIS-3FLAG pGAL-SCC1 SMC3-aid) were arrested in G1 in raffinose-containing medium with alpha factor for 3 hr before addition of 0.5 mM NAA. After 30 min, while maintaining G1 arrest, 2% galactose was added, and cultures were harvested for Scc2 and Scc1 ChIP after 45 min.
(E and F) Ectopic loading of cohesin in G1 is dependent on Chl4. Analysis of Scc1 and Scc2 loading at centromeres in G1-arrested wild-type and chl4Δ mutant cells, following ectopic expression of Scc1. Strains AM9334 (SCC2-6HIS-3FLAG pGAL-SCC1-3HA) and AM9613 (SCC2-6HIS-3FLAG, pGAL-SCC1-3HA chl4Δ) were treated as in (A) and (B), and qPCR analysis of ChIP samples (E) and immunoblot (F) are shown.
(G) Scc1 production enables Scc2 association with kinetochores throughout the cell cycle. Images are shown of live wild-type (AM10715) and chl4Δ cells (AM10716) carrying pGAL-SCC1-3HA, SCC2-GFP, and MTW1-tdTomato after culturing in galactose-containing medium for 2 hr.
(H) Model for the spatial and temporal control of cohesin loading by Scc1 and the Ctf19 complex.
In (A), (D), and (E) the mean of three independent experiments is shown with error bars representing standard error (∗∗p < 0.01; ∗p < 0.05, two-tailed paired t test). See also Figure S4 and Table S3.