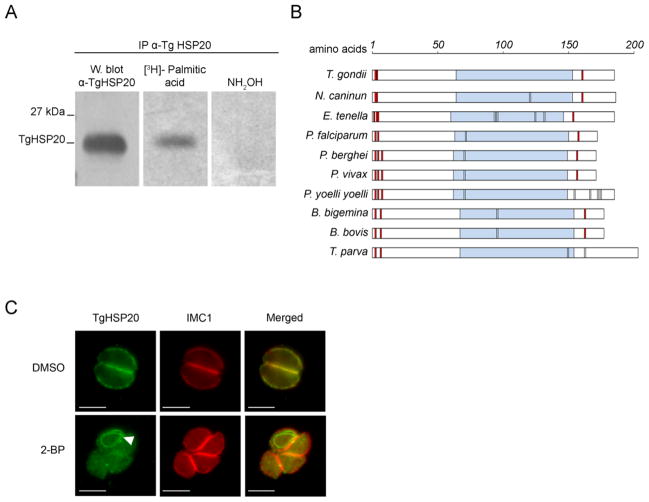
Figure 1. TgHSP20 is palmitoylated in vivo and this modification is responsible for its IMC localization.
A) TgHSP20 is palmitoylated in vivo. Metabolic labeling of wild type T. gondii parasites using [3H]-palmitic acid followed by specific immunoprecipitation with anti-TgHSP20 antibody. After the immunoprecipitation, the sample was divided in three and run on SDS-PAGE: one sample was used for western blotting as a loading control. The two remaining samples were used to detect the radiolabeled HSP20 as indicated in Materials and Methods. B) Schematic representation of Apicomplexan HSP20 amino acid sequences. The analysis includes the amino acid sequences of HSP20 from Toxoplasma gondii (TgHSP20; NCBI accession number AAT66039), Neospora caninum (NcHSP20; NCBI accession number CD537778), Eimeria tenella (EtHSP20; NCBI accession number AI758032), Plasmodium falciparum (PfHSP20; NCBI accession number CAD51208), Plasmodium berghei (PbHSP20; NCBI accession number AAL07529), Plasmodium vivax (PvHSP20; NCBI accession number XP_001615006), Plasmodium yoelii yoelii (PyyHSP20; NCBI accession number EAA16935), Babesia bigemina (BbiHSP20; NCBI accession number AAK11630), Babesia bovis (BbiHSP20; NCBI accession number AAK11624) and Theileria parva (TpHSP20; NCBI accession number XP_763804). Light-blue boxes indicate α-crystallin domain. Red boxes represent cysteine residues predicted to be palmitoylated. Black lines depict cysteine residues. C) TgHSP20 depends on palmitoylation for IMC localization. Intracellular parasites were treated for 16–18 h with either 100 μM 2-bromopalmitate (2-BP) or DMSO and then TgHSP20 localization was analyzed by indirect immunofluorescence. The localization of the protein IMC1 was unaffected by the treatment. An unknown structure stained with TgHSP20 antibodies is shown in cells treated with 2-BP (arrowhead). Scale bars = 5 μm.