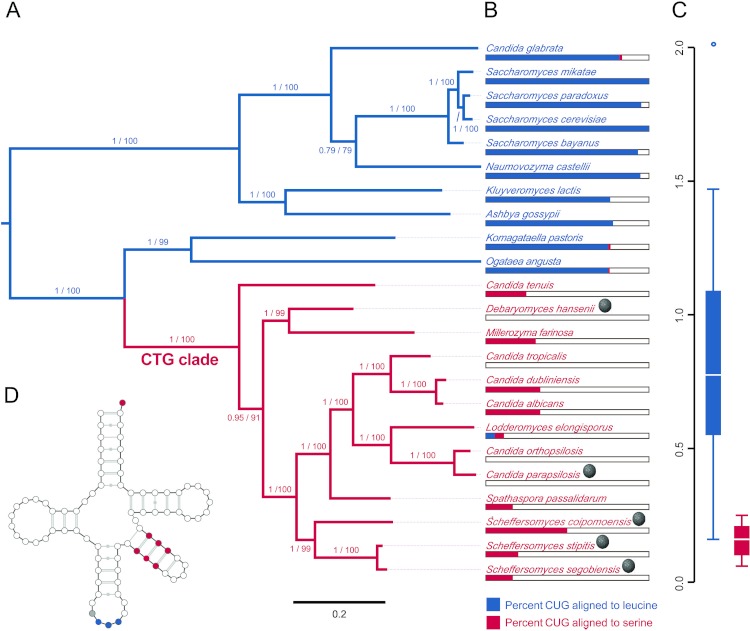
Figure 3. Evidence that the viral host, Scheffersomyces segobiensis, uses the modified genetic code of the “CTG” clade.
(A) Midpoint-rooted maximum likelihood phylogram of CTG clade yeasts and related yeasts based on the protein sequences of the five most phylogenetically informative genes for fungi. Branches are labeled with support values from approximate likelihood ratio tests and nonparametric bootstrapping. Blue shading indicates standard code yeasts, while red shading indicates CTG code yeasts. Gray spheres indicate lineages with evidence of past or prior infections with totiviruses. (B) Comparison of CUG codon positions in each taxon versus homologous amino acid residues in Saccharyomyces cerevisiae. Blue bars represent the percent of S. cerevisiae leucine residues that are coded by the CUG codon for each taxon, while red bars represent the percent of S. cerevisiae serine residues that are coded by CUG for each taxon. (C) Boxplots showing relative synonymous codon usage (RSCU) for CUG codon usage by each taxon, the blue plot represents CUG RSCU values for non-CTG clade yeast, while the red plot represents CTG clade yeast including S. segobiensis. (D) The secondary structure model of the CTG clade type of tRNASER detected in S. segobiensis. The red shading indicates serine identifier sites, while blue shading indicates standard leucine identifier sites. The gray site is a typical guanine residue of the tRNASER of CTG yeast that lowers the leucine amino-acylation efficiency.