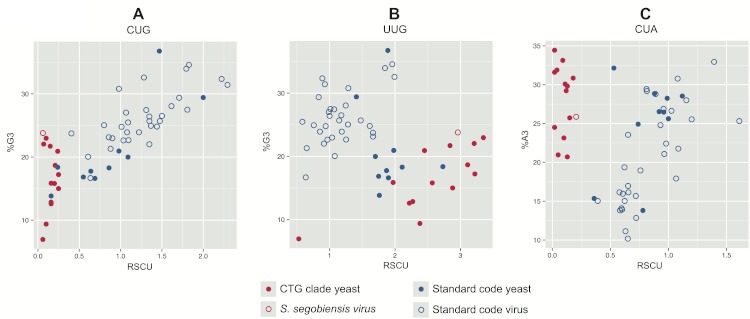
Figure 4. Bivariate plots of relative synonymous codon usage (RSCU) for serine and leucine versus third position base composition in yeast and their dsRNA viruses. CTG clade yeasts are shown as solid red points, and their viruses as hollow red points.
Standard code yeasts are shown as solid blue points and their viruses as hollow blue points. (A) CUG is used by standard code yeasts and their viruses but avoided by CTG clade yeasts and Scheffersomyces segobiensis virus L. (B) Leucine codon UUG is overused by CTG clade yeasts and S. segobiensis virus L relative to standard code yeasts and their viruses. (C) Leucine codon CUA is used by standard code yeasts and their viruses, but avoided by CTG clade yeasts and Scheffersomyces segobiensis virus L. Comparison with %N3 suggests that these codon preferences are not attributable to nucleotide composition alone.