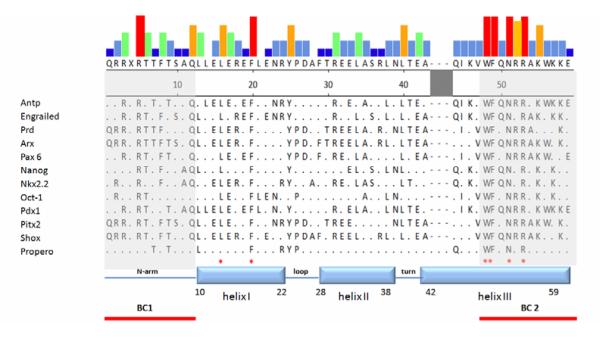
Figure 2. Sequences of homeodomains.
Selected homeoproteins were aligned by the Clustal W program [137] embedded in MegAlign (DNASTAR, Inc). The colors of the top panel represent the frequency of given residues among sequences shown. The red column represents perfectly conserved residues for which there are no exceptions. The yellow and green columns indicate conserved residues with exceptions (frequency around 70%-90%). The grey and blue columns indicate residues that are much less well conserved among the homeoproteins (from high to low: red>yellow>green>grey>blue). Four amino acid residues in helix III and two amino acid residues in helix I indicated by ‘*’ are conserved among all homeoproteins [32, 34, 49]. Note that two basic clusters are located at both ends of the homeodomain. The shaded region between helix II and helix III can form an extended loop for specific types of homeodomains.