Abstract
Objectives
To utilize high throughput techniques to analyze intestinal microbial ecology in premature neonates, who are highly susceptible to perturbations of the luminal environment associated with necrotizing enterocolitis (NEC) and late onset sepsis..
Study design
Using non-culture based techniques we evaluated intestinal microbiota shortly after birth and during hospitalization in 23 neonates born at 23-32 weeks gestational age. Microbiota compositions were compared in six preterm infants who developed NEC and/or signs of systemic inflammation versus matched controls using 16S rRNA pyrosequencing.
Results
Microbial DNA was detected in meconium suggesting an intrauterine origin. Differences in diversity were detected in infants whose mothers intended to breast feed (p=0·03), babies born to mothers with chorioamnionitis (p=0·06) and in babies born at less than 30 weeks gestation (p=0·03). A 16S rRNA sequence analysis detected Citrobacter-like sequences only in cases with NEC (three out of four) as well as an increased frequency of Enterococcus-like sequences in cases and Klebsiella in controls (p=0·06). The overall microbiota profiles in cases with NEC were not distinguishable from that in controls.
Conclusions
Microbial DNA in meconium of premature infants suggests prenatal influences. Microbial ecology alterations prior to NEC and sepsis suggest intestinal microbial origins.
Keywords: preterm infants, necrotizing enterocolitis, systemic inflammation, intestinal microbiota, bacteria
The human adult harbors an estimated 1×1014 cells, ~10% belong to the host and the remaining 90% represent the microbiota in or on the host, most of which are found in the intestinal tract.1-4 Although little data exist from human studies, animal studies suggest that the intestinal microbiota during the neonatal period has a profound effect on nutritional status, gastrointestinal (GI) tract development, and maintenance of mucosal surface integrity.2,5-7 Our understanding of the establishment and evolution of the intestinal microbiota during early infancy in humans has been limited because classical cultivation techniques allow the identification of a limited number of bacteria. In the adult human gut, 60-80% of the total microbiota can not be cultivated.8,9
In premature infants, the GI environment and susceptibility to disease are different compared with that in term infants.10 There is little information pertaining to the development of the intestinal microbiota using non-culture based techniques in infants. A few studies have monitored the bacterial communities in preterm infants.11-15 Application of high throughput 16S rRNA based techniques is needed to accurately assess the developing GI microbiota. 16S rRNA is part of the small ribosomal subunit that is preserved among all living organisms. It is the most universally used molecule for phylogenetic analysis as the gene encoding it contains conserved as well as variable regions. Similarity in the 16S rRNA genes is a measure of the relatedness of a bacterium of interest to other bacteria. Advantages of phylogenetic analysis using a 16S rRNA approach include its high sensitivity and the ability to analyze organisms without a need to first culture them. 454 pyrosequencing allows for an in depth analysis of 16S rRNA gene sequences derived from community DNA, such as that present in human feces. The large number of sequences obtained using the 454 pyrosequencing approach, which exceeds conventional Sanger based sequencing by magnitudes, permits the detection of abundant but also rare bacteria.
This research is critical because these infants have several immaturities of the GI tract,16-18 including hyperactive inflammatory responses19 to microbial antigens that may predispose the premature infant to intestinal injury including necrotizing enterocolitis (NEC) and intestine-derived systemic inflammation.7 This study was designed to use non-culture based technology to evaluate intra- and interindividual variation in early intestinal microbial ecology in premature infants to help guide investigations in larger, more extensive studies.
Methods
The study protocol was approved by the ethics committee at the University of Florida. Informed consent was obtained from the parents. The study protocol involved all the babies less than 32 weeks gestational age at birth. Parents of three infants refused to participate in the study. The first stool sample (meconium) and weekly samples thereafter were collected from 27 preterm infants, born at 23 to 32 weeks of gestational age, who had been admitted to the Neonatal Intensive Care Unit. Four of these babies’ samples did not have denaturing gradient gel electrophoresis (DGGE) profiling but were used only for pyrosequencing. For monitoring the GI microbiota profile, DGGE was performed on multiple fecal samples from 23 infants that were born at gestational ages that ranged from 23-32 weeks with birth weight ranging from 520g to 1997g (Table I).
Table I.
Clinical Characteristics of infants
| Characteristic | DGGE Group (n=23) frequency or mean (SD) |
|---|---|
| Male/Female | 15/8 |
| Gestational Age (weeks) | 29.9 (2.5) |
| Birth Weight (grams) | 1394 (420) |
| Total Number of Stools | 5.2 (3.3) |
| Delivery: C-section/Vaginal | 13/10 |
| Feeding: Breast Milk/Formula‡ | 15/8 |
| Infant: Antibiotics Y/N† | 19/4 |
| Mother: Antibiotics Y/N* | 7/16 |
| Prolonged Rupture of Membranes |
Y/N 6/17 |
intent to breast milk feed and actual breast milk feed group vs Formula
infants antibiotic therapy immediate after birth
Mother’s intrapartum antibiotic therapy
In order to evaluate in greater depth the intestinal microbial composition in infants with i) NEC and ii) presumptive sepsis with signs of systemic inflammatory response, we performed 454-based 16S rRNA pyrosequencing in a subset of 12 infants. Prospective stool samples were chosen approximately one week (range three to nine days) before onset of the disease from four subjects who developed NEC and two additional subjects who showed signs of sepsis and systemic inflammation (elevated C-reactive protein (CRP>10), elevated white blood cell counts and I/T ratio > 20, or culture positive sepsis). Six controls were matched by birth weight, week of delivery and the day of life of stool collection to each of the cases (Table II). All fecal samples were frozen at −80°C immediately after collection until analyses were performed.
Table II.
16S rRNA analysis prior the development inflammatory conditions
| Subject | Status | Gestational Age (weeks) |
Birth Weight (grams) |
Sex | Delivery Mode‡ |
Diet† | DOL* | Number of Sequences |
|---|---|---|---|---|---|---|---|---|
| B1 | NEC | 28 | 1000 | M | CS | B | 12 | 794 |
| B2 | Control | 32 | 1437 | M | CS | B/F | 13 | 1002 |
|
| ||||||||
| C1 | NEC | 29 | 1148 | M | CS | F | 24 | 830 |
| C2 | Control | 29 | 1134 | F | CS | F | 22 | 971 |
|
| ||||||||
| D1 | NEC | 31 | 1711 | M | CS | F | 4 | 755 |
| D2 | Control | 31 | 1395 | M | CS | F | 4 | 825 |
|
| ||||||||
| E1 | NEC | 29 | 1496 | M | VD | B/F | 13 | 963 |
| E2 | Control | 31 | 1377 | M | CS | F | 12 | 605 |
|
| ||||||||
| F1 | SIRS | 24 | 583 | M | VD | B | 24 | 881 |
| F2 | Control | 28 | 1026 | M | VD | F | 20 | 933 |
|
| ||||||||
| G1 | SIRS | 23 | 520 | F | VD | B | 58 | 1012 |
| G2 | Control | 27 | 705 | M | CS | F | 58 | 1085 |
CS=C-section
VD=Vaginal Delivery
B=actual breast milk feed
F-formula
DOL=Day of life of the stool sample collection.
Microbiota analysis
DNA extraction and DGGE
DNA was extracted from 200-300mg fecal samples using a modified Qiagen stool DNA extraction protocol.20 Primers were used to amplify the V6-V8 region as described by Zoetendal et al21 DGGE was performed in an 8% (wt/vol) polyacrylamide gel with a denaturing gradient ranging from 40% to 50% at the top and bottom of the gel, respectively (100% denaturing conditions were defined as 7M urea and 40% formamide). After electrophoresis (16h, 65V, 60°C), the gels were stained with SYBER Gold (Novex, San Diego, CA), scanned with Quantity One, and analyzed with Diversity Database software (Bio-Rad, Hercules, CA).
Estimation of microbial richness and diversity
The richness and diversity of the preterm infants’ fecal microbiota was determined from the number and intensity of PCR-DGGE bands present in each profile. Profiles for various time points were compared for each infant and between different infants. We calculated Simpson diversity index (SDI)22 and compared microbial diversity using 1/SDI for individual time points as well as the mean for each infant.
454 based 16S rRNA pyrosequencing
DNA from the fecal samples was amplified using a barcoded pyrosequencing primer based on universal primers 27F and 338R as described by Hamady et al23 Sequences of low quality or with a length of less than 150 nucleotides were removed from the analysis. Sequences were analyzed using the RDP pyrosequencing pipeline24 including features to calculate diversity indices and rarefaction curves. We used weighted UNIFRAC to generate a principal component analysis to compare overall similarity in microbiota composition.25
Statistics
The associations of prognostic factors (such as breast or formula feeding) with the Simpson diversity Index (SDI) were evaluated by the two-sample two-sided t-test. Where repeated measures were used, we calculated a personal mean of the SDI and compared the mean of the means between the two groups. Descriptive data are expressed as mean± S.D. In addition to the mean of mean analyses, which weigh all subjects equally, we conducted secondary nested repeated (mixed random effects) measures ANOVAs. The two give basically the same qualitative conclusions, and therefore only the mean of mean analyses are reported in detail. Trends over time were assessed by first obtaining a personal slope, and comparing these with a median of zero by a two-sided Wilcoxon sign-rank test. Nonparametric methods were used here as personal slopes tend to be outlier prone.
Student t-test was used to test the difference between two independent groups or samples and to determine the significance of differences in the number of specific OTU’s (operational taxonomic unit) that were detected in the sequence analysis. P< 0·05 was considered statistically significant. Similarity matrices for comparing overall microbiota composition were generated using weighted as well as unweighted UNIFRAC analysis, and they were further analyzed by principal component analysis.25
Results
Over a period of approximately one year, 27 preterm infants were enrolled (Table I). The time of first stool collection varied from second day of life to day 22 of life, one infant born at 23 weeks gestational age had a delayed passage of meconium on day of life 22. Depending on the length of hospital stay the number of stool samples collected per infant varied from one to 15.
Early Fecal Microbiota
In order to determine whether microbes were present in the meconium and to evaluate diversity depending on pre- and perinatal factors, we compared microbiota diversity in all initial fecal samples using DGGE profiling. For two of the 23 very premature infants we did not detect DNA in the first stool sample, they were therefore not included in this analysis. Microbiota diversity in the samples was compared using the SDI. Table III (left column) shows differences in diversity index from stool microbiota from the first stool samples from 21 babies. Lower gestational age (<30 compared with ≥ 30 weeks gestation) was associated with lower diversity index (p=0·03). Trends toward lower diversity index were evident in babies whose mothers had prolonged rupture of membranes (PROM) and who received intrapartum antibiotics (p=0·09 and p=0·06, respectively). Mothers’ intent to breast feed was associated with a lower diversity index (p=0·03). No significant differences in microbiota diversity were seen in babies of mothers who delivered by Cesarean section versus vaginal delivery (p=0·5).
Table III.
Simpson Diversity Index in Meconium and > 7 Days Postnatal Age
| Variable | Simpson Diversity Index Mean (SD) <n> |
||
|---|---|---|---|
| Meconium | >7 Days Postnatal Age |
||
|
| |||
| Maternal intrapartum Antibiotics |
Yes | 6.76 (2.90) <6> | 9.27 (2.52) <7> |
| No | 9.37 (2.55) <15> | 8.97 (1.73) <15> | |
| P-Value | 0.06 | 0.75 | |
|
| |||
| Feeding type | Breast Milk | ‡7.60 (2.58) <13> | †9.09 (2.03) <14> |
| Formula | 10.28 (2.60) <8> | 9.04 (2.13) <7> | |
| P-Value | 0.03 | 0.96 | |
|
| |||
| Gestational Age |
⋝30 | 9.38 (2.54) <16> | 9.31 (1.78) <15> |
| <30 | 6.20 (2.62) <5> | 8.56 (2.37) <7> | |
| P-Value | 0.03 | 0.42 | |
|
| |||
| Delivery | C-section | 8.99 (3.33) <12> | 9.64 (1.86) <12> |
| Vaginal | 8.13 (2.14) <9> | 8.39 (1.95) <10> | |
| P-Value | 0.50 | 0.14 | |
|
| |||
| Prolonged Rupture of Membranes |
Yes | 7.11 (2.80) <7> | 8.90 (2.25) <7> |
| No | 9.38 (2.65) <14> | 9·15 (1.89) <15> | |
| P-Value | 0.09 | 0.79 | |
intent to breast milk feed
type of breast milk feed
Diversity over time under different conditions
Of the 23 infants initially included, one was transferred to another hospital after the first stool collection and was not available for longitudinal microbiota profiling. Microbiota profiles determined in the remaining 22 infants by DGGE revealed unique compositions for each infant. Although composition changed over time, characteristic bands were observed for each infant at multiple time points. There was a trend towards increased diversity over time (data not shown). After removing samples collected during the first week of life from the analysis, the association between microbiota diversity and type of feeding breast milk (the infants who were in the intent to breast feed group remained in breast fed group) vs formula disappeared (Table III, right column). No significant associations were detected between microbiota diversity and sex, birth weight, gestational age (p=0·42), PROM (p=0·79), maternal intrapartum antibiotics (p=0·75), or mode of delivery (p=0·14)
454 pyrosequencing: NEC and/or suspected sepsis with signs of inflammation
In addition to microbiota profiling by DGGE, we performed an in depth 16S rRNA analysis for a 6 infants with NEC and/or suspected sepsis with signs of inflammation (cases) and six matched samples without signs of NEC or sepsis (controls) (Table II). Fecal samples were collected prospectively and 16S rRNA sequence analysis was performed on samples collected approximately one week (range three to nine days) before onset of symptoms. For the 12 samples we obtained a total of 13377 16S rRNA sequences with an average length of 244 nucleotides (Min = 46, Max = 319). We used the RDP pyrosequencing pipeline to remove low quality and short reads, retaining a total of 10656 reads (average of 888 sequences/sample). The number of OTU’s in each of the samples at the level of 100%, 99% and 97% identity ranged from 107-260; 48-116 and 18-46 respectively.
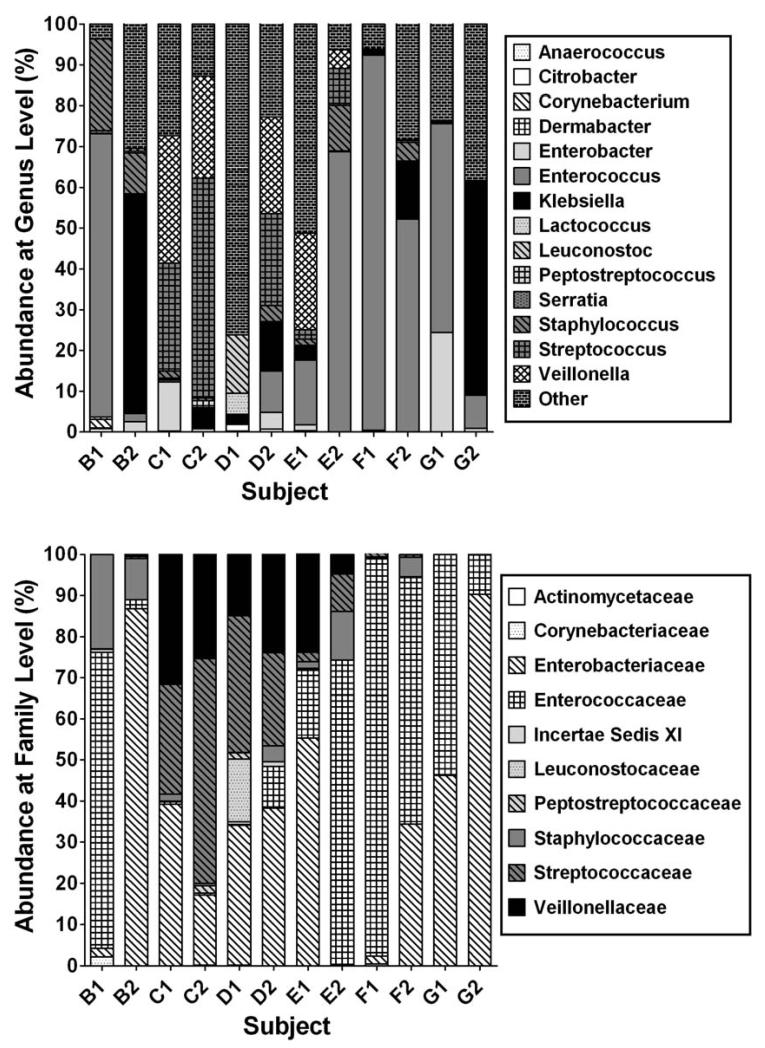
Differences were obvious at the level of bacterial family and were more pronounced at the level of bacterial genus (Figure; for the subject characteristics refer to Table II). When OTU’s were grouped by bacterial family ten families represented at least 98 % of all sequences observed (Figure). Each sample varied in the proportion of OTU’s belonging to a particular bacterial family. When OTU’s were grouped by genus level some similarities could be observed in cases and controls respectively (Figure). Higher numbers of Enterococcus were more frequently detected in cases with NEC and SIRS (systemic inflammatory response syndrome) compared with controls. In contrast, Klebsiella was more frequently detected in controls compared with both cases of NEC and SIRS (p=0·06). In addition we detected two OTUs with closest matches to Enterobacteriaceae that were detected in higher proportion in controls (p<0·05). Sequences matching Citrobacter species were detected, although present in low numbers, in 3 out of 4 cases of NEC but in none of the controls.
Figure.
Abundance of bacterial families & genera from Pyrosequencing Analysis Proportion of OTUs matching ten most frequently observed families. B1, C1, D1 and E1 represent cases with NEC, F1 and G1 SIRS, matched with controls B2, C2, D2, E2, F2, G2. Sequences matching other bacterial families represent < 2% in any of the samples are not shown. Subject characteristics and # of sequences are shown in Table II. “Other” represents minor and unclassified OTUs.
When overall microbiota composition was evaluated using a weighted Unifrac analysis,25 the resulting principal component analyses did not indicate that microbiota composition in cases is more similar than in controls. Cases and controls did not cluster into distinct groups as would be expected if overall microbiota composition was more similar within cases or controls.
Discussion
The goal of this study was to gain insight into the establishment of intestinal microbiota composition in premature infants born under 32 weeks of gestational age. We investigated changes in microbiota over time to evaluate potential associations with environmental factors such as gestational age, mode of delivery, premature rupture of membranes, use of maternal antibiotics, and breast versus formula feeding. In addition, we generated initial data on potential contributions of distortions in microbial ecology to the onset of the NEC or late onset sepsis. Our findings, based on non-culture based techniques, show that microbiota in preterm infants shows a high inter-individual variability that develops surprisingly rapidly after birth. In the earliest stool samples, diversity differed depending on gestational age at delivery, use of maternal antibiotics, prolonged rupture of membranes, and intended feeding type (breast versus formula). No significant differences were detected in these early samples between C-section versus vaginally delivered infants. Comparison of infants stool microbiota prior to the development of inflammatory processes such as NEC or sepsis using 454 pyrosequencing suggested differences in colonization patterns. One of the findings using both DGGE and 454 pyrosequencing is the marked variability of microbial composition between infants. However, within individual babies, several bands persisted over time with a trend toward increasing diversity.
Millar et al26 was the first to use 16S rRNA gene PCR and evaluated the bowel flora of preterm infants with and without NEC. Fecal samples from 32 preterm infants, including samples from 10 infants with NEC, were examined by culture and PCR amplification of the 16S rRNA gene. In this study, uncultured bacteria detected by PCR-DGGE were no more frequent in the fecal samples of infants with NEC than in the samples of infants without NEC. Previous studies that utilized non-culture based techniques agree partially with our findings. Magne et al13 also found a high inter-individual variability. The main groups encountered were the Enterobacteria family and the genera Enterococcus, Streptococcus, and Staphylococcus. Our study is in agreement with the finding of high inter-individual variability in the bands found on the gels and also 454 pyrosequencing, but in addition shows a rapid establishment of diversity, which after the first samples is not markedly different when comparisons are made under several environmental conditions.
In samples collected before onset of disease symptoms and analyzed by high throughput 16S rRNA sequencing we detected correlations between the presence/absence of specific bacterial groups and future disease status. Each preterm infant developed a microbiota composition that was clearly distinct from that in others, even in the samples collected from twins. Another study using a combination of culture and molecular methods for analyzing bifidobacterial colonization12 showed that colonization by bifidobacteria was affected neither by birth weight, mode of delivery, antibiotics given to the mother, and infants nor type of feeding, but gestational age at birth was a significant condition for colonization by bifidobacteria. However, in contrast to our study, which longitudinally evaluated infants at 23-32 weeks gestational age, this study didn’t involve premature babies born at less than 30 weeks. These observations are suggestive of differences in the proportion of specific bacterial groups that should be evaluated in larger studies.
De la Cochetiere et al27 previously reported the prospective detection of a temperature gradient gel electrophoresis (TGGE) band that corresponded to C. perfringens in 3 cases of NEC and none of 9 controls. We did not detect any particular band shared between cases only. Although we sequenced more than 10K 16S rRNA fragments, very few sequences had their closest match to the genus Clostridia and no sequences matched C. perfringens. Thus, our results do not support an association between C. perfringens and NEC.
We evaluated microbial DNA in the first stools of premature babies using non-culture based methods. It is likely that the microbes from the meconium of these infants are not of postnatal origin. Previous studies have demonstrated the presence of microbes in amniotic fluid without rupture of membranes using both culture and non culture based techniques.28,29 In one of these, a correlation was found between extent of microbial colonization and length of gestation.28 Our finding suggests that amniotic fluid containing microbes may have been swallowed in utero. The lower microbial diversity in the infants with gestational age < 30 weeks gestation appears to contradict the findings of DiGiulio,27 If increasing degree of prematurity is associated with more amniotic fluid microbial colonization, then one would also expect to find more microbes in the first stools of the more premature infants. Additional studies are needed to further evaluate this apparent discrepancy.
Previous studies using primarily culture-based techniques have suggested differences in microbial colonization of breast versus formula fed infants, but this remains controversial.30 Of interest in our study is the lower diversity of the first stools from babies of mothers who intended to breast feed. Most of these babies had received little or no feeding at the time of the first stool collection. Whether this is due to differences in antenatal care, socioeconomic differences, or other factors can only be speculated upon at this time.
We also found a trend toward less diversity in the first stools of babies whose mothers received antibiotics. This difference diminished over time. The effect of postnatal antibiotics could not be determined in this study because the majority of these premature babies received antibiotics in the first several days of postnatal life. Routine use of antibiotics in the premature has recently been associated with a higher likelihood of developing NEC.31
In conclusion, these data provide an important foundation for future studies. These include evaluations of the very early microbiota in premature infants with early onset of labor, the effects of pre- and postnatal antibiotics on the early intestinal microbial ecology and correlations with clinical effects and the effects of various nutrients and feeding regimens on the intestinal microbial environment and subsequent development of disease.
Acknowledgement
We thank M. Hamady and R. Knight for help with the UNIFRAC analysis.
This work was partly supported by the National Institute of Research Resources, National Institutes of Health; grant numbers: RO1 HD 059143 and M01RR00082 and an educational grant to M. Mshvildadze from the European Society for Pediatric Research.
Abbreviations
- NEC
necrotizing enterocolitis
- GI
gastrointestinal
- DGGE
denaturant gradient gel electrophoresis
- SDI
Simpson diversity index
- OTU
operational taxonomic unit
- SIR
systemic inflammatory response
- PROM
prolonged rupture of membranes
- SIRS
systemic inflammatory response syndrome
- CRP
C-reactive protein
- MAB
maternal antibiotics
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare no conflicts of interest.
References
- 1.Hattori M, Taylor TD. The Human Intestinal Microbiome: A New Frontier of Human Biology. DNA Res. 2009:dsn033. doi: 10.1093/dnares/dsn033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dethlefsen L, Eckburg PB, Bik EM, Relman DA. Assembly of the human intestinal microbiota. Trends Ecol Evol. 2006;21(9):517–23. doi: 10.1016/j.tree.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 3.Dethlefsen L, McFall-Ngai M, Relman DA. An ecological and evolutionary perspective on human-microbe mutualism and disease. Nature. 2007;449(7164):811–818. doi: 10.1038/nature06245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. The human microbiome project. Nature. 2007;449(7164):804–10. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hooper LV, Gordon JI. Commensal Host-Bacterial Relationships in the Gut. Science. 2001;292:1115–1118. doi: 10.1126/science.1058709. [DOI] [PubMed] [Google Scholar]
- 6.Hooper LV. Bacterial contributions to mammalian gut development. Trends Microbiol. 2004;12(3):129–34. doi: 10.1016/j.tim.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 7.Caicedo RA, Schanler RJ, Li N, Neu J. The developing intestinal ecosystem: implications for the neonate. Pediatr Res. 2005;58(4):625–8. doi: 10.1203/01.PDR.0000180533.09295.84. [DOI] [PubMed] [Google Scholar]
- 8.Riesenfeld CS, Schloss PD, Handelsman J. Metagenomics: genomic analysis of microbial communities. Annu Rev Genet. 2004;38(38):525–52. doi: 10.1146/annurev.genet.38.072902.091216. [DOI] [PubMed] [Google Scholar]
- 9.Frank DN, Pace NR. Gastrointestinal microbiology enters the metagenomics era. Curr Opin Gastroenterol. 2008;24(1):4–10. doi: 10.1097/MOG.0b013e3282f2b0e8. [DOI] [PubMed] [Google Scholar]
- 10.Claud EC, Walker WA. Hypothesis: inappropriate colonization of the premature intestine can cause neonatal necrotizing enterocolitis. FASEB J. 2001;15(8):1398–403. doi: 10.1096/fj.00-0833hyp. [DOI] [PubMed] [Google Scholar]
- 11.Gewolb IH, Schwalbe RS, Taciak VL, Harrison TS, Panigrahi P. Stool microflora in extremely low birthweight infants. Arch Dis Child Fetal Neonatal Ed. 1999;80(3):F167–73. doi: 10.1136/fn.80.3.f167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Butel MJ, Suau A, Campeotto F, Magne F, Aires J, Ferraris L, et al. Conditions of bifidobacterial colonization in preterm infants: a prospective analysis. J Pediatr Gastroenterol Nutr. 2007;44(5):577–82. doi: 10.1097/MPG.0b013e3180406b20. [DOI] [PubMed] [Google Scholar]
- 13.Magne F, Abély M, Boyer F, Morville P, Pochart P, Suau A. Low species diversity and high interindividual variability in faeces of preterm infants as revealed by sequences of 16S rRNA genes and PCR-temporal temperature gradient gel electrophoresis profiles. FEMS Microbiol Ecol. 2006;57(1):128–38. doi: 10.1111/j.1574-6941.2006.00097.x. [DOI] [PubMed] [Google Scholar]
- 14.Mshvildadze M, Neu J, Mai V. Intestinal microbiota development in the premature neonate: establishment of a lasting commensal relationship? Nutr Rev. 2008;66(11):658–63. doi: 10.1111/j.1753-4887.2008.00119.x. [DOI] [PubMed] [Google Scholar]
- 15.Neu J, Douglas-Escobar M, Lopez M. Microbes and the Developing Gastrointestinal Tract. NCP - Nutrition in Clinical Practice. 2007;22(2):174–182. doi: 10.1177/0115426507022002174. [DOI] [PubMed] [Google Scholar]
- 16.Neu J. Functional development of the fetal gastrointestinal tract. Semin. Perinatol. 1989;13(3):224–35. [PubMed] [Google Scholar]
- 17.Neu J. Gastrointestinal development and meeting the nutritional needs of premature infants. Am J Clin Nutr. 2007;85(2):629S–634. doi: 10.1093/ajcn/85.2.629S. [DOI] [PubMed] [Google Scholar]
- 18.Neu J, Mackey AD. Neonatal Gastrointestinal Innate Immunity. NeoReviews. 2003;4:14–18. [Google Scholar]
- 19.Nanthakumar NN, Fusunyan RD, Sanderson I, Walker WA. Inflammation in the developing human intestine: A possible pathophysiologic contribution to necrotizing enterocolitis. Proc Natl Acad Sci U S A. 2000;97(11):6043–6048. doi: 10.1073/pnas.97.11.6043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mai V, Greenwald B, Morris JG, Jr., R JP, Stine OC. Effect of bowel preparation and colonoscopy on post-procedure intestinal microbiota composition. Gut. 2006;55:1822–1823. doi: 10.1136/gut.2006.108266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zoetendal EG, Ben-Amor K, Akkermans AD, Abee T, de Vos WM. DNA isolation protocols affect the detection limit of PCR approaches of bacteria in samples from the human gastrointestinal tract. Syst Appl Microbiol. 2001;24(3):405–10. doi: 10.1078/0723-2020-00060. [DOI] [PubMed] [Google Scholar]
- 22.Begon M, Harper JL, R TC. Ecology: Individuals, Populations, and Communities. 3rd edition Blackwell Science Ltd.; Cambridge, MA: 1996. [Google Scholar]
- 23.Hamady M, Walker JJ, Harris JK, Gold NJ, Knight R. Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. Nat Methods. 2008;5(3):235–7. doi: 10.1038/nmeth.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71(12):8228–35. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lozupone C, Hamady M, Knight R. UniFrac--an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics. 2006;7:371. doi: 10.1186/1471-2105-7-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Millar MR, Linton CJ, Cade A, Glancy D, Hall M, Jalal H. Application of 16S rRNA gene PCR to study bowel flora of preterm infants with and without necrotizing enterocolitis. J Clin Microbiol. 1996;34:2506–2510. doi: 10.1128/jcm.34.10.2506-2510.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de la Cochetiere MF, Piloquet H, des Robert C, Darmaun D, Galmiche JP, Roze JC. Early intestinal bacterial colonization and necrotizing enterocolitis in premature infants: the putative role of Clostridium. Pediatr Res. 2004;56(3):366–70. doi: 10.1203/01.PDR.0000134251.45878.D5. [DOI] [PubMed] [Google Scholar]
- 28.DiGiulio DB, Romero R, Amogan HP, Kusanovic JP, Bik EM, Gotsch F, et al. Microbial prevalence, diversity and abundance in amniotic fluid during preterm labor: a molecular and culture-based investigation. PLoS ONE. 2008;3(8):33056. doi: 10.1371/journal.pone.0003056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goldenberg RL, Culhane JF, Iams JD, Romero R. Epidemiology and causes of preterm birth. Lancet. 2008;371(9606):75–84. doi: 10.1016/S0140-6736(08)60074-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fanaro S, Chierici R, Guerrini P, Vigi V. Intestinal microflora in early infancy: composition and development. Acta Paediatr Suppl. 2003 Sep;91(441):48–55. doi: 10.1111/j.1651-2227.2003.tb00646.x. 2003;91(441):48-55. [DOI] [PubMed] [Google Scholar]
- 31.Cotten CM, Taylor S, Stoll B, Goldberg RN, Hansen NI, Sánchez PJ, et al. Prolonged duration of initial empirical antibiotic treatment is associated with increased rates of necrotizing enterocolitis and death for extremely low birth weight infants. Pediatrics. 2009;123(1):58–66. doi: 10.1542/peds.2007-3423. [DOI] [PMC free article] [PubMed] [Google Scholar]