Table 3. Predicted annotations of mouse genes for erythrocyte development (GO:0048821).
| MGI ID | Gene | Existing GO annotation | Allele supporting new annotation |
| MGI:1921354 | Abcb6 |
Abcb6
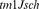
|
|
| MGI:2158492 | Ahsp | GO:0030218 erythrocyte differentiation | |
| MGI:88064 | Ar | ArTfm | |
| MGI:88113 | Atp4a |
Atp4a
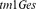
|
|
| MGI:106190 | Bcl11a |
Bcl11a

|
|
| MGI:1338013 | Cbfa2t3 |
Cbfa2t3
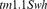
|
|
| MGI:88316 | Ccne1 |
Ccne1
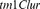
|
|
| MGI:96617 | Cd47 |
Cd47
|
|
| MGI:103198 | Cdc25a |
Cdc25a

|
|
| MGI:2384875 | Cdk5rap2 |
Cdk5rap2

|
|
| MGI:1344352 | Dido1 |
Dido1

|
|
| MGI:1329019 | Dnase2a | GO:0030218 erythrocyte differentiation | |
| MGI:1330300 | Dyrk3 | GO:0030218 erythrocyte differentiation | |
| MGI:103012 | E2f4 |
E2f4
|
|
| MGI:95295 | Egr1 |
Egr1
|
|
| MGI:95401 | Epb4.1 |
Epb4.1
|
|
| MGI:95407 | Epo | GO:0030218 erythrocyte differentiation | |
| MGI:95408 | Epor |
Epor
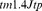
|
|
| MGI:109336 | Etv6 |
Etv6

|
|
| MGI:1351611 | Exoc6 | GO:0030218 erythrocyte differentiation | |
| MGI:95513 | Fech | GO:0030218 erythrocyte differentiation | |
| MGI:95661 | Gata1 | GO:0048821 erythrocyte development | |
| MGI:95662 | Gata2 | GO:0045648 positive regulation of erythrocyte differentiation | |
| MGI:1276578 | Gfi1b | GO:0045646 regulation of erythrocyte differentiation | |
| MGI:96103 | Hk1 |
Hk1
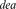
|
|
| MGI:96109 | Hlx |
Hlx
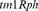
|
|
| MGI:1098219 | Hmgb3 | GO:0045638 negative regulation of myeloid cell differentiation | |
| MGI:96187 | Hoxb6 | GO:0034101 erythrocyte homeostasis | |
| MGI:1342540 | Ikzf1 |
Ikzf1
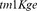
|
|
| MGI:96560 | Il6st |
Il6st
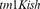
|
|
| MGI:107357 | Inpp5d | GO:0045648 positive regulation of erythrocyte differentiation | |
| MGI:96395 | Irf8 | GO:0030099 myeloid cell differentiation | |
| MGI:96629 | Jak2 | GO:0030218 erythrocyte differentiation | |
| MGI:99928 | Jak3 |
Jak3
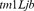
|
|
| MGI:104813 | Jarid2 |
Jarid2
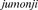
|
|
| MGI:96677 | Kit | GO:0030218 erythrocyte differentiation | |
| MGI:96974 | Kitl |
Kitl
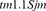
|
|
| MGI:1354948 | Klf13 | GO:0045647 negative regulation of erythrocyte differentiation | |
| MGI:96757 | Lcn2 |
Lcn2
|
|
| MGI:96785 | Lhx2 |
Lhx2
|
|
| MGI:101789 | Lig1 |
Lig1
|
|
| MGI:1346865 | Mapk14 | GO:0045648 positive regulation of erythrocyte differentiation | |
| MGI:2444881 | Mfsd7b | GO:0030218 erythrocyte differentiation | |
| MGI:3619412 | Mir451 |
Mir451
|
|
| MGI:97249 | Myb |
Myb
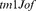
|
|
| MGI:2442415 | Myst3 | GO:0030099 myeloid cell differentiation | |
| MGI:1349717 | Ncor1 |
Ncor1

|
|
| MGI:104741 | Nfkbia | GO:0045638 negative regulation of myeloid cell differentiation | Nfkbiatm1.1Kbp |
| MGI:97566 | Pgm3 |
Pgm3
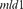
|
|
| MGI:2385902 | Picalm |
Picalm
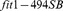
|
|
| MGI:97583 | Pik3r1 |
Pik3r1

|
|
| MGI:1201409 | Pknox1 | GO:0030218 erythrocyte differentiation | |
| MGI:101898 | Pou2f1 |
Pou2f1
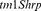
|
|
| MGI:1927072 | Ppp1r15a |
Ppp1r15a
|
|
| MGI:109486 | Prdx2 |
Prdx2
|
|
| MGI:97806 | Ptpn2 |
Ptpn2

|
|
| MGI:97810 | ptprc |
Ptprc
|
|
| MGI:97874 | Rb1 | GO:0043353 enucleate erythrocyte differentiation | |
| MGI:103289 | Relb |
Relb
|
|
| MGI:99852 | Runx1 | GO:0030099 myeloid cell differentiation |
Runx1
|
| MGI:2146974 | Safb |
Safb

|
|
| MGI:2445054 | Senp1 | GO:0010724 regulation of definitive erythrocyte differentiation | |
| MGI:98282 | Sfpi1 | GO:0045646 regulation of erythrocyte differentiation | |
| MGI:1928761 | slc19a2 |
Slc19a2
|
|
| MGI:108392 | Slc20a1 |
Slc20a1
|
|
| MGI:98354 | Sos1 |
Sos1
|
|
| MGI:1277166 | Sp3 | GO:0030218 erythrocyte differentiation | Sp3tm1Sus |
| MGI:98385 | Spna1 |
Spna1
|
|
| MGI:1915678 | Steap3 |
Steap3

|
|
| MGI:98480 | Tal1 | GO:0045648 positive regulation of erythrocyte differentiation | |
| MGI:1196624 | Tcea1 | GO:0030218 erythrocyte differentiation | |
| MGI:98822 | Tfrc |
Tfrc
|
|
| MGI:98729 | Tgfbr2 |
Tgfbr2

|
|
| MGI:1920999 | Ttc7 |
Ttc7
|
|
| MGI:1270126 | Ulk1 |
Ulk1
|
|
| MGI:98917 | Uros |
Uros
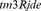
|
|
| MGI:103223 | Vhl |
Vhl
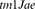
|
|
| MGI:1095400 | Zfpm1 | GO:0060318 definitive erythrocyte differentiation |
Figure 1 shows the part of the GO hierarchy containing erythrocyte development. Of the 77 genes predicted by phenotypic analysis to annotate to erythrocyte development, 29 already had some relevant annotation, shown in column 3. Relevant annotation was taken to be any child class of myeloid cell differentiation (GO:0030099), or erythrocyte homeostasis (GO:0034101), thereby including as many levels of granularity as possible in order to compensate for possible curator decisions to annotate more generally. The remaining 48 genes had no existing annotations to any of these classes. In many cases, multiple genotypes provided evidence for the novel annotation; an example allele is shown in column 4. Phenotype annotations to abnormal erythropoiesis (MP:0000245) or its subclasses were counted as evidence. Whilst close curation of the phenotypic evidence may suggest that annotation to a parent of erythrocyte development is more appropriate, in all cases the evidence indicated that annotation to the neighbourhood of this class was correct but missing.
