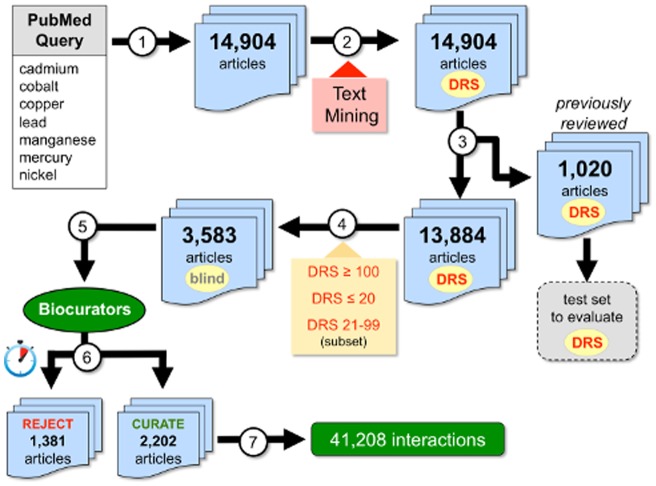
Figure 2. Document workflow.
(1) Independent CTD-specific queries were made of PubMed to retrieve 14,904 articles for the seven heavy metals cadmium, cobalt, copper, lead, manganese, mercury, and nickel. (2) These articles were text mined and assigned a document relevancy score (DRS). (3) Of this preliminary corpus, 1,020 articles were found to have been previously reviewed in CTD and were used as a test set to evaluate the DRS and determine suitable cut-offs. (4) Articles with DRS ≥100 (high), DRS ≤20 (low), and a subset with DRS between 21–99 (medium) were combined to provide a final corpus of 3,583 documents which was then (5) sent to five CTD biocurators (who were kept blind to the DRS of each article) for review. (6) Biocurators timed themselves while reviewing all articles and ultimately rejected 1,381 (as non-curatable for CTD) and curated 2,202 of them (7) from whence 41,208 chemical-gene-disease interactions were extracted.