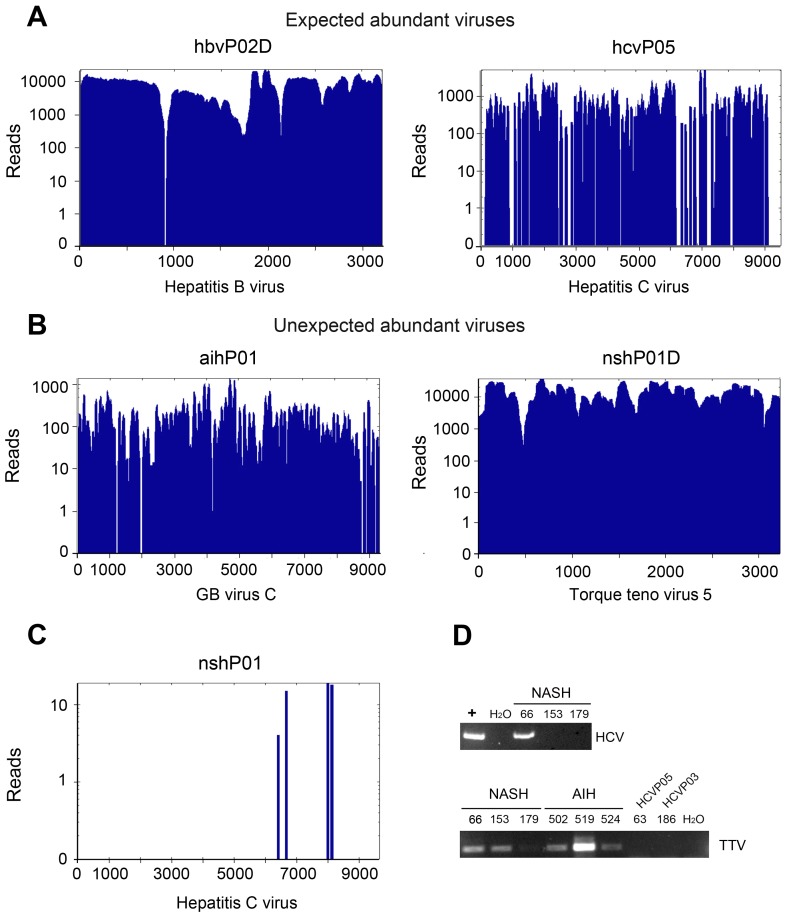
Figure 4. NGS identified several classes of viruses that would otherwise remain undetected.
Panels A to C show the mapping of reads along the indicated viral genome, (A) Expected abundant viruses were recovered at high and uniform coverage as illustrated for HBV (NC_003977.1) in library hbvP02D and HCV genotype 2 (NC_009823.1) in library hcvP05. (B) Unexpected abundant viruses were also found at high and uniform coverage, as illustrated for GBV-C/Hepatitis G virus (NC_001710.1) in library aihP01 and TTV 5 isolate TCHN-C1 (AF345523.1) in library nshP01D. (C) A few read ends mapping to HCV genotype 1 (NC_004102.1) were found in library nshP01, which initially tested negative for HCV in serological tests. (D) HCV was PCR amplified in one of the three samples pooled for construction of library nshP01 (upper panel). Additionally, TTV was found in samples NSH and AIH, but not in samples from patients affected by chronic hepatitis C (HCVP03 and HCVP05).