Figure 1.
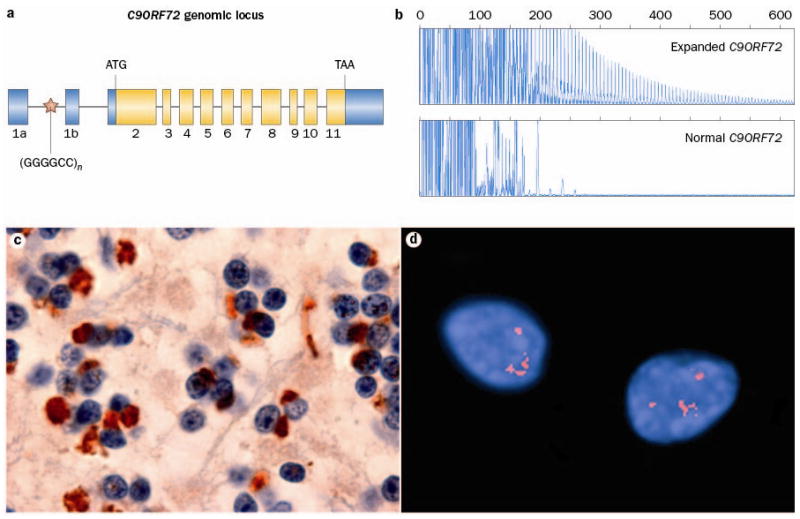
Expanded GGGGCC hexanucleotide repeat in non-coding region of C9ORF72 causes FTD and ALS linked to chromosome 9p. a Overview of the genomic structure of C9ORF72. Numbered boxes represent coding (white) and non-coding (gray) exons and the position of the start codon (ATG) and stop codon (TAA) are indicated. The position of the (GGGGCC)n repeat in the intronic region between exons 1a and 1b is indicated with a red star. b PCR products of repeat-primed PCR reactions separated on an ABI3730 DNA Analyzer and visualized by GENEMAPPER software. Electropherograms are zoomed to 2,000 relative fluorescence units to show stutter amplification. One FTD patient with a pathogenic expanded C9ORF72 repeat (top) and one FTD patient with a C9ORF72 normal repeat length (bottom) are shown. c In addition to FTLD-TDP and ALS pathology, all patients with the C9ORF72 mutation show a unique pattern of ubiquitin-positive (brown), TDP-43-negative neuronal inclusions in the cerebellar granule layer and other specific neuroanatomical regions. This disease-specific finding implies the mis-metabolism and accumulation of some yet unidentified protein(s). d RNA foci, visualized using a Cy3-labeled (GGCCCC)4 oligonucleotide probe (red), in the nuclei of two lower motor neurons from an FTD-ALS patient carrying the expanded GGGGCC repeat in C9ORF72.
