Figure S1.
iCLIP Data Show Binding of hnRNP C and U2AF65 at 3′ Splice Sites of Target Exons, Related to Figure 1
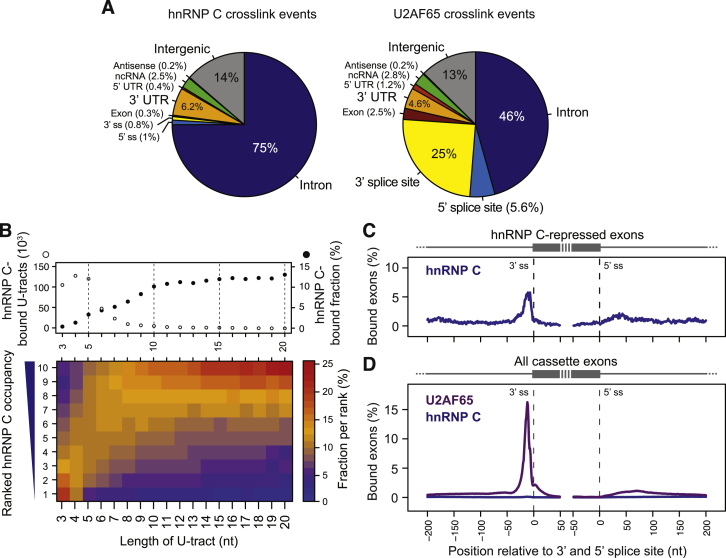
(A) The majority of hnRNP C and U2AF65 crosslink events is located within introns. Pie chart summarizing the fraction of crosslink events within different genomic regions. 3‘ splice site (3′ ss) and 5‘ splice site (5′ ss) indicate the regions 200 nt upstream and downstream of Ensembl-annotated exons, respectively.
(B) Binding sites on longer U-tracts show higher hnRNP C occupancy. Top panel: absolute numbers of hnRNP C-bound U-tracts of different length in the transcriptome (open circles) and the corresponding fraction of bound tracts (filled circles). Lower panel: heatmap showing the fraction of U-tracts of a given length that is allocated to the ten different ranks of hnRNP C occupancy. 3-nt U-tracts show the highest enrichment at the 10% weakest hnRNP-C-binding sites (rank 1), whereas U-tracts of nine or more nucleotides show the highest enrichment at the 10% strongest binding sites (rank 10).
(C) hnRNP C binding is enriched immediately upstream of the 3′ splice sites of hnRNP-C-repressed exons. RNA map showing the percentage of exons that have a crosslink nucleotide at a certain position relative to the 3′ and 5′ splice site of all exons that are repressed by hnRNP C (taken from our RNA-seq data, Table S2).
(D) U2AF65 binding is strongly enriched immediately upstream of the 3′ splice sites of all annotated exons. RNA map as in (C) visualizing U2AF65 and hnRNP C iCLIP data relative to all 3′ and 5′ splice sites annotated in the Ensembl database.