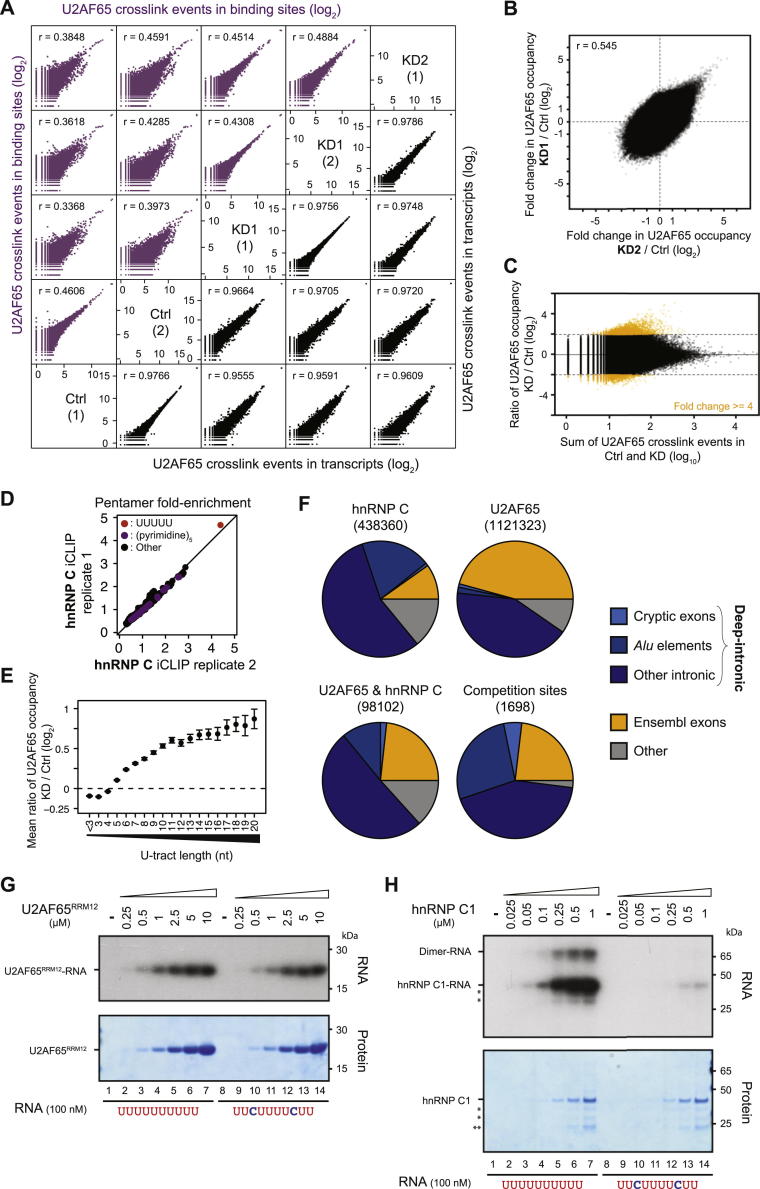
Figure S3.
hnRNP C Competes with U2AF65 Binding at Long U-Tracts and Cryptic Exons, Related to Figure 2
(A) Scatter plots comparing the U2AF65 iCLIP crosslink events within binding sites (purple) or Ensembl transcripts (black) for all replicate experiments from control HeLa cells (Ctrl) and HNRNPC knockdown cells (KD1 and KD2). Sample types and replicate numbers (in brackets) are given along the diagonal. The Spearman’s rank correlation (r) for each pair is given in the upper left corner of the respective panel.
(B) The changes in U2AF65 occupancies on individual binding sites are highly correlated between both knockdowns. Scatter plot comparing the fold changes in U2AF65 occupancy in the two different HNRNPC knockdowns (KD1 and KD2). The occupancy was calculated by normalizing the number of crosslink events within each binding site by the total number of crosslink events within the corresponding gene. The Pearson’s product-moment correlation (r) is given in the upper left corner.
(C) U2AF65 shows significantly changed occupancy on a large number of binding sites in the HNRNPC knockdown. Plot showing the ratio of U2AF65 occupancy from the combined HNRNPC knockdowns (KD) over Ctrl against the sum of U2AF65 crosslink events in all samples. The 5,500 binding sites that show an at least 4-fold change in U2AF65 occupancy are shown in yellow. The dashed lines mark a 4-fold change in either direction.
(D) hnRNP C preferentially crosslinks to UUUUU pentamers. Scatter plot comparing the pentamer fold-enrichment at crosslink sites from two hnRNP C iCLIP replicate experiments. hnRNP C prefers the pentamer UUUUU (red), but shows no particular enrichment for further pyrimidine combinations (purple) or other pentamers (black).
(E) U2AF65-binding sites that overlap with a long U-tract show increased U2AF65 occupancies in the HNRNPC knockdown. The average ratio of U2AF65 occupancy from HNRNPC knockdown (KD) over control HeLa cells (Ctrl) is shown for U2AF65-binding sites that overlap with U-tracts of varying lengths. Error bars indicated the 95% confidence interval of the mean.
(F) Sites of hnRNP C-U2AF65 competition are strongly enriched at deep-intronic locations (p value < 0.001 compared with all other U2AF65-binding sites, hypergeometric test), in particular at cryptic exons and within Alu elements. Pie charts showing the fraction of binding sites located at annotated Ensembl exons as well as at deep-intronic positions (subdivided into positions at cryptic exons, within Alu elements and other). Graphs are shown from left to right for: all hnRNP C, all U2AF65 binding sites, U2AF65 binding sites that overlap with hnRNP C binding, and U2AF65 binding sites that show an at least 4-fold increase in occupancy in the HNRNPC knockdown and overlap with hnRNP C (competition sites).
(G) Recombinant U2AF65RRM12 shows comparable crosslinking to the wild-type (U10) and mutant (U2CU4CU2) RNA oligonucleotides that resemble the upstream U-tract of the Alu exon in the NUP133 minigene (Figure S6). Increasing concentrations of U2AF65RRM12 (21 kDa, concentration indicated above in μM) were incubated with radioactively labeled wild-type or mutant RNA oligonucleotide (100 nM), UV crosslinked and analyzed by denaturing gel electrophoresis. The radioactive signal from the RNA crosslinked to the protein can be observed in the autoradiograph (U2AF65RRM12-RNA, top panel). Coomassie staining of the same gel serves as loading control (lower panel).
(H) Recombinant hnRNP C1 crosslinking is drastically reduced to the mutant RNA oligonucleotide. Experimental setup as in (G) but using increasing concentrations of hnRNP C1 (33 kDa, concentration indicated above). Note that in addition to the signal derived from hnRNP C1 crosslinked to RNA (hnRNP C1-RNA), an additional crosslinking signal is visible at about 65 kDa. This signal is likely derived from two hnRNP C1 proteins crosslinked to one RNA molecule and therefore labeled as dimer-RNA. Impurities are indicated by asterisks on the left (∗ C-terminal truncations of hnRNP C1; ∗∗ GST).