Figure S4.
RNA-Seq Identifies a Large Number of hnRNP-C-Repressed Alu Exons, Related to Figure 3
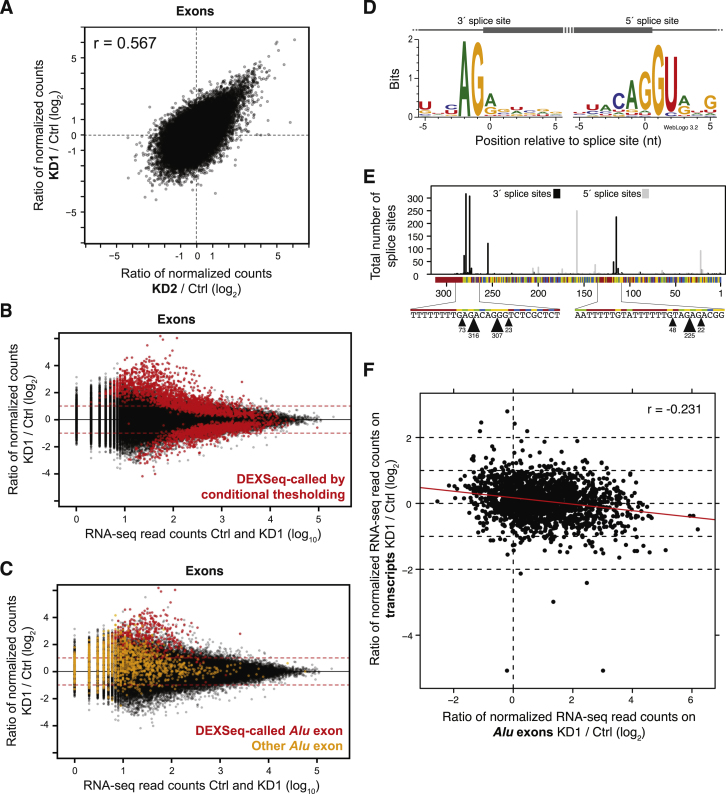
(A) The detected splicing changes show a strong correlation between both HNRNPC knockdowns. Scatter plot comparing the fold changes (log2) in normalized exon expression (HNRNPC knockdown over control HeLa cells) from both HNRNPC knockdowns (KD1 and KD2). The Pearson’s product-moment correlation (r) is given in the upper left corner.
(B) The majority of changed exons show higher inclusion in the HNRNPC knockdown. Plot indicating the fold change (log2) in normalized exon expression in KD1 against the sum of RNA-seq reads detecting the exon in KD1 and control samples. Exons that are called by DEXSeq are indicated in red (p values used for conditional thresholding pa = 0.01 and pb = 0.05).
(C) The Alu exons show widespread upregulation in the HNRNPC knockdown. Plot as in (B). DEXSeq-called Alu exons are highlighted in red, and all other Alu exons are depicted in yellow.
(D) The Alu exons arise from canonical splice sites. Weblogos indicating the consensus sequence at the 3′ and 5′ splice sites of the Alu exons.
(E) Bar diagram summarizing the usage of different 3′ (black) and 5′ (gray) splice sites in the Alu consensus sequence (color coding as for enlarged sequence below). The region around the most commonly used 3′ splice sites is enlarged below (black arrowheads; the number of exons using each splice site is given below the arrowhead).
(F) Scatter plot comparing changes in Alu exon inclusion (x axis) with changes in the corresponding transcript levels. The linear regression line (red) indicates the negative correlation between both data sets (Pearson’s product-moment correlation r = −0.231).