Figure S5.
Polypyrimidine Tracts of Established Bona Fide Alu Exons Exhibit Shorter Contiguous U-Tracts, Related to Figures 3 and 4
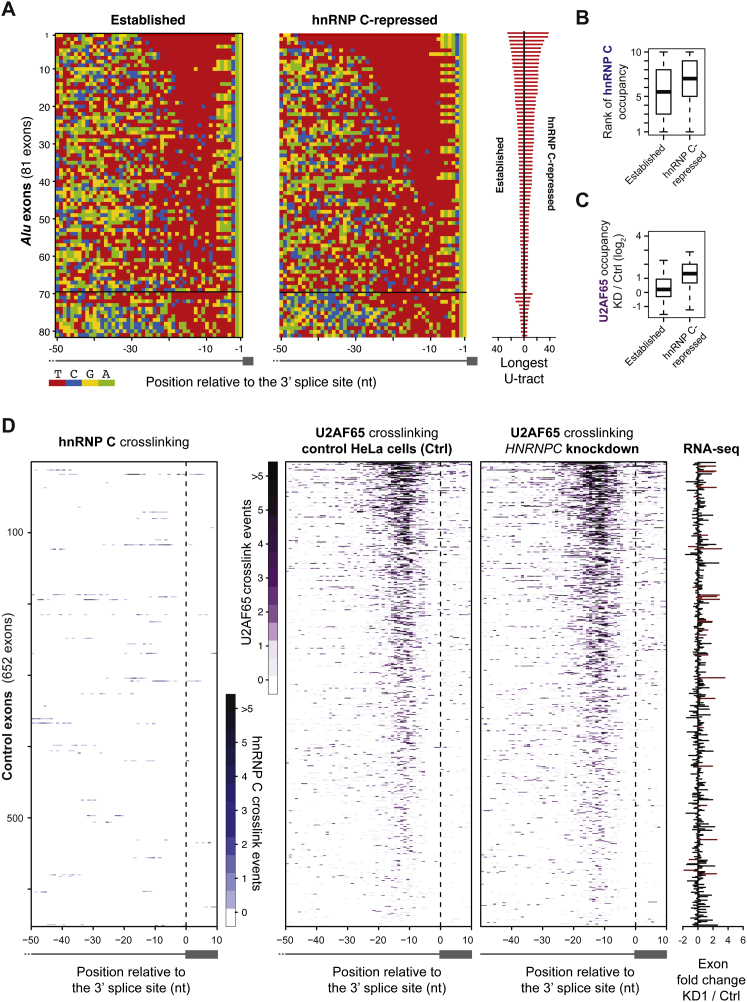
(A) Established Alu exons accumulate point mutations that disrupt the long U-tracts. Heatmaps visualizing the polypyrimidine tract of Alu exons originating from the first (top) and second (bottom) arm of Alu elements (separated by a horizontal black line). Each line represents the polypyrimidine tract of an individual Alu exon, with the different colors indicating the four bases (T: red; C: blue; G: yellow; A: green). In the left panel, the 81 pyrimidine tracts of established bona fide Alu exons are shown that are highly included and show a less than 1.5-fold change in either direction, while the right panel shows the same regions of a randomly selected subset of hnRNP-C-repressed Alu exons. The exons were sorted according to the longest contiguous U-tract, as indicated in the bar chart on the right.
(B) Established Alu exons harbor weaker hnRNP-C-binding sites. Box plot comparing the distribution of ranks of hnRNP-C-binding sites in front of 81 established or 81 randomly selected hnRNP-C-repressed Alu exons (p value < 0.1, Student’s t test).
(C) U2AF65-binding sites at established Alu exons show hardly any change in occupancy upon knockdown of HNRNPC. Box plot depicting the ratio of U2AF65 occupancies from HNRNPC knockdown (KD) and control HeLa cells (Ctrl) of U2AF65-binding sites in front of 81 established or 81 randomly selected Alu exons (p value < 10−9, Student’s t test).
(D) Control exons show little hnRNP C crosslinking and no increase in U2AF65 crosslinking in the HNRNPC knockdown. Heatmaps comparing the crosslinking of hnRNP C and U2AF65 relative to the 3′ splice site of the non-Alu exons downstream of the Alu exons shown in Figure 4. Each row corresponds to one of 652 exons. The left panel shows the crosslink events of hnRNP C at each nucleotide (nt) depicted in different shades of blue as indicated aside. The middle and right panels visualize U2AF65 crosslinking (different shades of purple) in control HeLa (Ctrl) and HNRNPC knockdown cells (KD). The bar diagram on the right (RNA-seq) shows the corresponding fold change in exon inclusion (KD1 / WT). Fold change values found to be significant according to the conditional thresholding approach are shown in red.