Figure S6.
hnRNP C Suppresses the Exonization of an Alu Element in the NUP133 Gene through Competition with U2AF65, Related to Figure 5
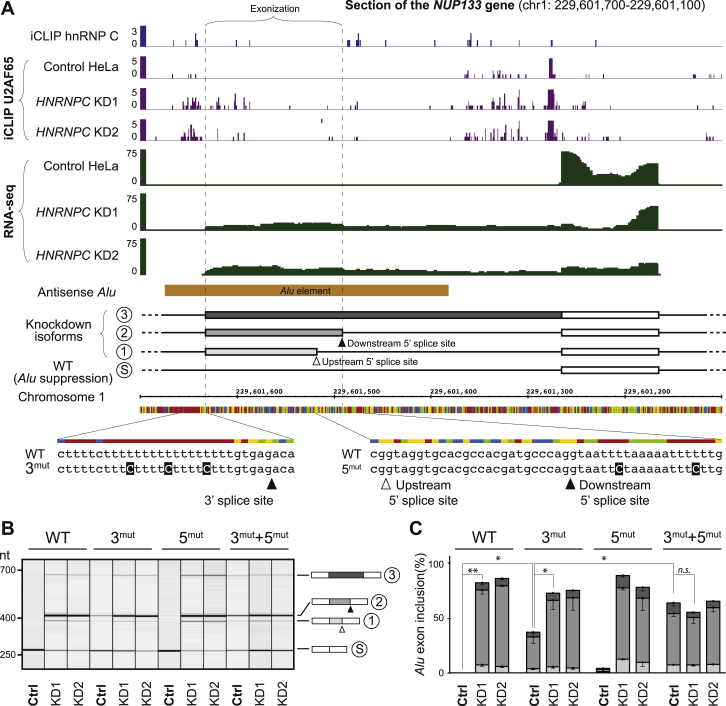
(A) Genome browser view including an Alu element (orange) plus the downstream exon within the NUP133 gene on chromosome 1 (229,601,700-229,601,100). hnRNP C iCLIP data (blue) show crosslinking at the upstream and the linker U-tract of the Alu element (corresponding thymidines are highlighted in red at the bottom). Little U2AF65 crosslinking (purple) is detected at the U-tracts in control HeLa cells, while strong crosslinking is observed in the HNRNPC knockdowns (KD1 and KD2). RNA-seq data (green) show exonization of the Alu element in both knockdowns, giving rise to three different Alu exon variants. The corresponding isoforms are schematically indicated: Alu suppression in isoform S, Alu inclusion as a cassette exon in isoform 1 and 2 (differing in the usage of an upstream or downstream 5′ splice site, respectively) and Alu inclusion using an alternative 3‘ splice site for the downstream exon (isoform 3). The wild-type (WT) sequence surrounding the 3′ and 5′ splice sites (arrowheads) including the introduced point mutations (3mut and 5mut) are shown below. The positions of the point mutations are highlighted in the sequences by black squares.
(B) U-to-C transitions promote Alu exonization in the presence of hnRNP C. Shown is the gel-like view of capillary electrophoresis of RT-PCR analysis of minigenes containing the Alu element in the NUP133 gene described in (A) with two flanking exons. The wild-type (WT) minigene shows no Alu exonization in control HeLa cells (Ctrl) and a significant increase in isoforms 1-3 in the HNRNPC knockdown (KD1 and KD2). A mutant minigene containing three T-to-C transitions in the upstream U-tract (3mut) shows significant inclusion of isoforms 1-3 in control HeLa cells. Additional T-to-C transitions in the linker U-tract (5mut) further elevate the inclusion of isoforms 1-3 in control HeLa cells and prevent any further regulation by hnRNP C (KD1 and KD2). The sizes corresponding to the different analyzed isoforms are schematically indicated on the right.
(C) Average Alu exon inclusion in percent for three replicate RT-PCR experiments as described in (B). The different isoforms are indicated by shades of gray (light gray: isoform 1; medium gray: isoform 2; dark gray: isoform 3). Lines indicate relevant comparisons with asterisks indicating different levels of significance for changes in the summed inclusion isoforms (∗: p value < 0.05; ∗∗: < 10−3; n.s.: not significant; Student’s t test). Error bars represent SD of the mean; n=3.