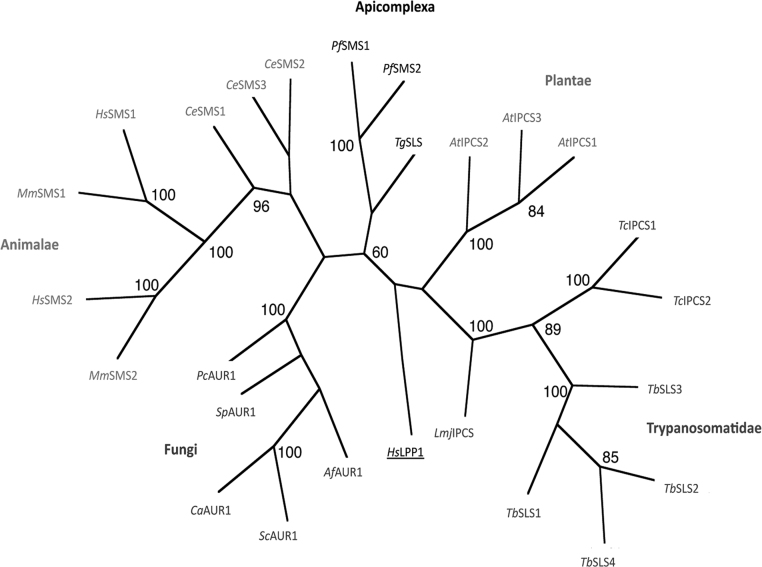
Supplementary Figure 1.
Maximum parsimony analyses of Animalae, Fungi, Trypanosomatidae, Plantae and Apicomplexa sphingolipid synthase predicted amino acid sequences. Bootstrap scores >60 indicated. Homo sapiens LPP1 (outgroup) accession number: O14494; T. gondii SLS: TGME49_046490; P. falciparum SMS1&2: PFF1210w and PFF1215w; Arabidopsis thaliana IPCS1-3: At3g54020.1, At2g37940.1, At2g29525.1; T. brucei SLS1-4: Tb09.211.1030, Tb09.211.1020, Tb09.211.1010, Tb09.211.1000; T. cruzi IPCS1&2: Tc00.1047053506885.124, Tc00.1047053510729.290; L. major IPCS: LmjF35.4990; Aspergillus fumigatus AUR1p: AAD22750; Candida albicans AUR1p: AAB67233; Pneumocystis carinii AUR1p: CAH17867; Saccharomyces cerevisiae AUR1p: NP_012922; Schizosaccharomyces pombe AUR1p: Q10142; Caenorhabditis elegans SMS1-3: Q9U3D4, AAA82341, AAK84597; Homo sapiens SMS1&2: AB154421, Q8NHU3; Mus musculus SMS1&2: Q8VCQ6, Q9D4B1.